Abstract
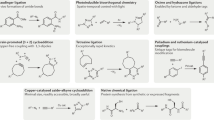
The 2022 Nobel Prize in Chemistry recognized the development of biorthogonal chemical ligation reactions known as click chemistry in biomedicine. This concept has catalyzed significant progress in sensing and diagnosis, chemical biology, materials chemistry, and drug discovery and delivery. In proteomics, the ability to incorporate a click tag into proteins has propelled development of powerful new methods for selective enrichment of protein complexes that inform understanding of protein networks. It also has had a strong influence on the ability to enrich for protein post-translational modifications. This feature article summarizes the impacts of biorthogonal click chemistry on proteomics.
Graphical Abstract




Similar content being viewed by others
References
Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–21.
Tornøe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67(9):3057–64.
Dong R, Yang X, Wang B, Ji X. Mutual leveraging of proximity effects and click chemistry in chemical biology. Medicinal Research Reviews.n/a(n/a):1–24.
Schreiber CL, Smith BD. Molecular conjugation using non-covalent click chemistry. Nat Rev Chem. 2019;3(6):393–400.
Wu D, Yang K, Zhang Z, Feng Y, Rao L, Chen X, et al. Metal-free bioorthogonal click chemistry in cancer theranostics. Chem Soc Rev. 2022;51(4): 1336–76.
Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276(5315):1125–8.
Saxon E, Bertozzi CR. Cell surface engineering by a modified staudinger reaction. Science. 2000;287(5460):2007–10.
Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430(7002):873–7.
Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide−alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126(46):15046–7.
Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320(5876):664–7.
Parker CG, Pratt MR. Click chemistry in proteomic investigations. Cell. 2020;180(4):605–32.
Smith LM, Kelleher NL. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10(3):186–7.
Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, et al. How many human proteoforms are there? Nat Chem Biol. 2018;14(3):206–14.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci USA. 2005;102(43):15545–50.
Hogrebe A, von Stechow L, Bekker-Jensen DB, Weinert BT, Kelstrup CD, Olsen JV. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat Commun. 2018;9(1):1045.
Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–3.
Adelmant G, Garg BK, Tavares M, Card JD, Marto JA. Tandem affinity purification and mass spectrometry (TAP-MS) for the analysis of protein complexes. Curr Protoc Protein Sci. 2019;96(1): e84.
Cho KF, Branon TC, Udeshi ND, Myers SA, Carr SA, Ting AY. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat Protoc. 2020;15(12):3971–99.
Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468(7325):790–5.
Zhao Q, Ouyang X, Wan X, Gajiwala KS, Kath JC, Jones LH, et al. Broad-spectrum kinase profiling in live cells with lysine-targeted sulfonyl fluoride probes. J Am Chem Soc. 2017;139(2):680–5.
Lapinsky DJ, Johnson DS. Recent developments and applications of clickable photoprobes in medicinal chemistry and chemical biology. Future Med Chem. 2015;7(16):2143–71.
Hart GW, Ball LE. Post-translational modifications: a major focus for the future of proteomics. Mol Cell Proteomics. 2013;12(12):3443.
Dennis JW. Genetic code asymmetry supports diversity through experimentation with posttranslational modifications. Curr Opin Chem Biol. 2017;41:1–11.
Zachara NE, Akimoto Y, Boyce M, Hart GW. The O-GlcNAc modification. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors. Essentials of Glycobiology. 4th ed. Cold Spring Harbor (NY)2022. p. 251–64.
Ma J, Wang WH, Li Z, Shabanowitz J, Hunt DF, Hart GW. O-GlcNAc site mapping by using a combination of chemoenzymatic labeling, copper-free click chemistry, reductive cleavage, and electron-transfer dissociation mass spectrometry. Anal Chem. 2019;91(4):2620–5.
Hinneburg H, Stavenhagen K, Schweiger-Hufnagel U, Pengelley S, Jabs W, Seeberger PH, et al. The art of destruction: optimizing collision energies in quadrupole-time of flight (Q-TOF) instruments for glycopeptide-based glycoproteomics. J Am Soc Mass Spectrom. 2016;27(3):507–19.
Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, Bertozzi CR. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat Methods. 2015;12(6):561–7.
Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131(13):4967–75.
Kulkarni RA, Worth AJ, Zengeya TT, Shrimp JH, Garlick JM, Roberts AM, et al. Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem Biol. 2017;24(2):231–42.
Islam K, Chen Y, Wu H, Bothwell IR, Blum GJ, Zeng H, et al. Defining efficient enzyme-cofactor pairs for bioorthogonal profiling of protein methylation. P Natl Acad Sci USA. 2013;110(42):16778–83.
Kalesh K, Lukauskas S, Borg AJ, Snijders AP, Ayyappan V, Leung AKL, et al. An integrated chemical proteomics approach for quantitative profiling of intracellular ADP-ribosylation. Sci Rep. 2019;9(1):6655.
Funding
The author is supported by the US National Institute for General Medical Sciences grant R35GM144090.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The author is an editor of Analytical and Bioanalytical Chemistry and did not participate in the review of this manuscript.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zaia, J. The 2022 Nobel Prize in Chemistry for the development of click chemistry and bioorthogonal chemistry. Anal Bioanal Chem 415, 527–532 (2023). https://doi.org/10.1007/s00216-022-04483-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04483-9