Abstract
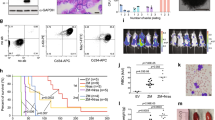
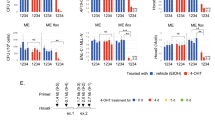
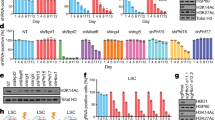
Histone acetyltransferases (HATs) catalyze the transfer of an acetyl group from acetyl-CoA to lysine residues of histones and play a central role in transcriptional regulation in diverse biological processes. Dysregulation of HAT activity can lead to human diseases including developmental disorders and cancer. Through genome-wide CRISPR-Cas9 screens, we identified several HATs of the MYST family as fitness genes for acute myeloid leukemia (AML). Here we investigate the essentiality of lysine acetyltransferase KAT7 in AMLs driven by the MLL-X gene fusions. We found that KAT7 loss leads to a rapid and complete loss of both H3K14ac and H4K12ac marks, in association with reduced proliferation, increased apoptosis, and differentiation of AML cells. Acetyltransferase activity of KAT7 is essential for the proliferation of these cells. Mechanistically, our data propose that acetylated histones provide a platform for the recruitment of MLL-fusion-associated adaptor proteins such as BRD4 and AF4 to gene promoters. Upon KAT7 loss, these factors together with RNA polymerase II rapidly dissociate from several MLL-fusion target genes that are essential for AML cell proliferation, including MEIS1, PBX3, and SENP6. Our findings reveal that KAT7 is a plausible therapeutic target for this poor prognosis AML subtype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq and ChIP-seq data are available from Gene Expression Omnibus under accession number GSE133516.
References
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
Saultz JN, Garzon R. Acute myeloid leukemia: a concise review. J Clin Med Res. 2016;5:33.
Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33.
Winters AC, Bernt KM. MLL-rearranged leukemias—an update on science and clinical approaches. Front Pediatr. 2017;5:4.
Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan W-I, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33.
Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8.
Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–95.
Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65.
Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–25.
Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–84.
Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016;17:1193–205.
Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168:890–903.e15.
Tzelepis K, De Braekeleer E, Aspris D, Barbieri I, Vijayabaskar MS, Liu W-H, et al. SRPK1 maintains acute myeloid leukemia through effects on isoform usage of epigenetic regulators including BRD4. Nat Commun. 2018;9:5378.
Sheikh BN, Akhtar A. The many lives of KATs—detectors, integrators and modulators of the cellular environment. Nat Rev Genet. 2019;20:7–23.
Voss AK, Thomas T. MYST family histone acetyltransferases take center stage in stem cells and development. Bioessays. 2009;31:1050–61.
Avvakumov N, Côté J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–407.
Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–33.
Kitabayashi I, Aikawa Y, Yokoyama A, Hosoda F, Nagai M, Kakazu N, et al. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia. 2001;15:89–94.
Panagopoulos I, Fioretos T, Isaksson M. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13). Mol Genet. 2001;10:395–404.
Esteyries S, Perot C, Adelaide J, Imbert M, Lagarde A, Pautas C, et al. NCOA3, a new fusion partner for MOZ/MYST3 in M5 acute myeloid leukemia. Leukemia. 2008;22:663–5.
Borrow J, Stanton VP Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41.
Valerio DG, Xu H, Chen C-W, Hoshii T, Eisold ME, Delaney C, et al. Histone acetyltransferase activity of MOF is required for MLL-AF9 leukemogenesis. Cancer Res. 2017;77:1753–62.
Lalonde M-E, Avvakumov N, Glass KC, Joncas F-H, Saksouk N, Holliday M, et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27:2009–24.
Mishima Y, Miyagi S, Saraya A, Negishi M, Endoh M, Endo TA, et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 2011;118:2443–53.
Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31:845–60.
Feng Y, Vlassis A, Roques C, Lalonde M-E, González-Aguilera C, Lambert J-P, et al. BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016;35:176–92.
Doyon Y, Cayrou C, Ullah M, Landry A-J, Côté V, Selleck W, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64.
Tarumoto Y, Lu B, Somerville TDD, Huang Y-H, Milazzo JP, Wu XS, et al. LKB1, salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol Cell. 2018;69:1017–1027.e6.
Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–7.
Wunderlich M, Mizukawa B, Chou F-S, Sexton C, Shrestha M, Saunthararajah Y, et al. AML cells are differentially sensitive to chemotherapy treatment in a human xenograft model. Blood. 2013;121:e90–7.
Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, et al. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem. 2008;283:28817–26.
Thorne RMW, Milne TA. Dangerous liaisons: cooperation between Pbx3, Meis1 and Hoxa9 in leukemia. Haematologica. 2015;100:850–3.
Chen M, Zhu N, Liu X, Laurent B, Tang Z, Eng R, et al. JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors. Genes Dev. 2015;29:2123–39.
Kerry J, Godfrey L, Repapi E, Tapia M, Blackledge NP, Ma H, et al. MLL-AF4 spreading identifies binding sites that are distinct from super-enhancers and that govern sensitivity to DOT1L inhibition in leukemia. Cell Rep. 2017;18:482–95.
Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–22.
Yokoyama A, Somervaille TCP, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–18.
Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46.
Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–21.
Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381:484–95.
Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78.
Voss AK, Collin C, Dixon MP, Thomas T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell. 2009;17:674–86.
Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–36.
Brookes E, Pombo A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 2009;10:1213–9.
Sauer T, Arteaga MF, Isken F, Rohde C, Hebestreit K, Mikesch J-H, et al. MYST2 acetyltransferase expression and Histone H4 Lysine acetylation are suppressed in AML. Exp Hematol. 2015;43:794–802.e4.
Lavallée V-P, Baccelli I, Krosl J, Wilhelm B, Barabé F, Gendron P, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47:1030–7.
MacPherson L, Anokye J, Yeung MM, Lam EYN, Chan Y-C, Weng C-F, et al. HBO1 is required for the maintenance of leukaemia stem cells. Nature. 2020;577:266–70.
Acknowledgements
This work was funded by the Wellcome Trust (WT206194), the Kay Kendall Leukaemia Fund (KKL920), Bloodwise (17006), Takeda Science Foundation, and Exonate Ltd. K.T. was funded by a Wellcome Trust Sir Henry Wellcome Fellowship (grant reference RG94424). G.S.V. was funded by a Cancer Research UK Senior Cancer Fellowship (C22324/A23015) and a Wellcome Trust Senior Fellowship in Clinical Science (WT095663MA). We thank Bee Ling Ng, Jennifer Graham, Christopher Hall, and Sam Thompson from the Wellcome Sanger Institute Cytometry Core Facility team for help with flow cytometry. We are grateful to the staff of the Sanger Institute Research Support Facility for help with mouse experiments and the staff of the Sanger Institute Core Sequencing facility for sequencing. We thank Mathew Garnett for providing the Nomo-1 cell line, Chris Vakoc for RN2 and MA9 cell lines, Pedro for his help compiling figures using Adobe Illustrate and Josep Nomdedeu for help advices in writing the manuscript.
Author information
Authors and Affiliations
Contributions
KY conceived the study and designed the experiments. YZA primarily performed the experiments and analyzed the data. M.Gu and SHO conducted bioinformatic analyses. EDB performed and analyzed the in vivo mouse studies. MG, EDB, and DA performed ChIP-qPCR and CRISPR validation studies. YT and JC performed cell proliferation assays. XC advised on ChIP experiments. JY, BJPH, MG, and KT helped with data interpretation and direction. YZA, MG, JY, GSV, and KY wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
GSV is a consultant for Kymab and Oxstem. All the remaining authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Au, Y.Z., Gu, M., De Braekeleer, E. et al. KAT7 is a genetic vulnerability of acute myeloid leukemias driven by MLL rearrangements. Leukemia 35, 1012–1022 (2021). https://doi.org/10.1038/s41375-020-1001-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-020-1001-z
This article is cited by
-
Chromatin complex dependencies reveal targeting opportunities in leukemia
Nature Communications (2023)
-
Histone acetylation by HBO1 (KAT7) activates Wnt/β-catenin signaling to promote leukemogenesis in B-cell acute lymphoblastic leukemia
Cell Death & Disease (2023)
-
MOZ/ENL complex is a recruiting factor of leukemic AF10 fusion proteins
Nature Communications (2023)
-
Integration of CRISPR/Cas9 with artificial intelligence for improved cancer therapeutics
Journal of Translational Medicine (2022)
-
NAA60 (HAT4): the newly discovered bi-functional Golgi member of the acetyltransferase family
Clinical Epigenetics (2022)