EGFR Expression in HER2-Driven Breast Cancer Cells
Abstract
:1. Introduction
2. Results
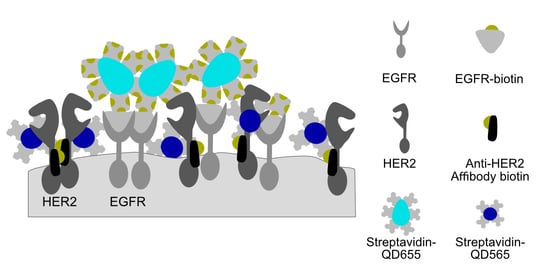
2.1. Establishment of a Dual-Labeling Approach for EGFR and HER2 for Correlative Light and Electron Microscopy
2.2. Growth Factor Expression Differs on Individual SKBR3 Cells
2.3. Large Membrane Protrusions Contain most HER2 Receptors
2.4. EGFR Expression on SKBR3 is Limited to a Subgroup and to Large Membrane Protrusions
3. Discussion
4. Materials and Methods
4.1. Materials and Chemicals Were Purchased from the Following Companies
4.2. Mammalian Cell Culture
4.3. Coating of Microchips and Cell Culture Microscope Dishes
4.4. Cell Seeding on Microchips or Cell Culture Microscope Dishes
4.5. Dual Labeling of EGFR and HER2, CD44 Immunofluorence Labeling
4.6. Differential Interference Contrast and Fluorescence Microscopy
4.7. Graphene Enclosure of Microchips for Electron Microscopy
4.8. Scanning Transmission Electron Microscopy (STEM)
4.9. Particle Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA | bovine serum albumin |
| CB | sodium cacodylate trihydrate buffer |
| DIC | differential interference contrast |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DPBS | Dulbecco’s PBS |
| EGF | epidermal growth factor |
| EGFR | EGF receptor |
| EGFR-QD655 | EGFR coupled to EGF conjugated biotin coupled to streptavidin QD655 |
| FBS | fetal bovine serum |
| FITC | Fluorescein isothiocyanate |
| GA | glutaraldehyde |
| GS | goat serum |
| HER2 | human epidermal growth factor receptor 2 |
| HER2-QD565 | HER2 bound to biotinylated anti-HER2 Affibody coupled to streptavidin QD565 |
| HPLC | high pressure liquid chromatography |
| LMP | large membrane protrusion |
| NEAAs | non-essential amino acids |
| PBS | phosphate buffered saline |
| PI3K | phosphoinositide 3-kinase |
| PLL | poly-L-lysine |
| QD | quantum dot |
| SF-DMEM | serum-free DMEM |
| STEM | scanning transmission electron microscopy |
| streptQD | streptavidin quantum dots |
References
- Hynes, N.E.; Lane, H.A. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.W.C.; Lie, E.F.; Toi, M. Advances in EGFR/HER2-directed clinical research on breast cancer. Adv. Cancer Res. 2020, 147, 375–428. [Google Scholar] [CrossRef]
- Hynes, N.E.; MacDonald, G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009, 21, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J.; Ferguson, K.M. The EGFR family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2014, 6, a020768. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.A.; Codony-Servat, J.; Albanell, J.; Rojo, F.; Arribas, J.; Baselga, J. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res. 2001, 61, 4744–4749. [Google Scholar]
- Badache, A.; Hynes, N.E. A new therapeutic antibody masks ErbB2 to its partners. Cancer Cell 2004, 5, 299–301. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, J.; Liu, C.; Du, S.; Feng, L.; Luan, X.; Zhang, Y.; Shi, Y.; Wang, T.; Wu, Y.; et al. Neratinib induces ErbB2 ubiquitylation and endocytic degradation via HSP90 dissociation in breast cancer cells. Cancer Lett. 2016, 382, 176–185. [Google Scholar] [CrossRef]
- Scaltriti, M.; Verma, C.; Guzman, M.; Jimenez, J.; Parra, J.L.; Pedersen, K.; Smith, D.J.; Landolfi, S.; Ramon y Cajal, S.; Arribas, J.; et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene 2009, 28, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Liu, Q.; Han, X.; Qin, S.; Zhao, W.; Li, A.; Wu, K. Development and clinical application of anti-HER2 monoclonal and bispecific antibodies for cancer treatment. Exp. Hematol. Oncol. 2017, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Guo, P.; Pu, T.J.; Chen, S.A.; Qiu, Y.; Zhong, X.R.; Zheng, H.; Chen, L.; Bu, H.; Ye, F. Breast cancers with EGFR and HER2 co-amplification favor distant metastasis and poor clinical outcome. Oncol. Lett. 2017, 14, 6562–6570. [Google Scholar] [CrossRef] [Green Version]
- Ingthorsson, S.; Andersen, K.; Hilmarsdottir, B.; Maelandsmo, G.M.; Magnusson, M.K.; Gudjonsson, T. HER2 induced EMT and tumorigenicity in breast epithelial progenitor cells is inhibited by coexpression of EGFR. Oncogene 2016, 35, 4244–4255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoadley, K.A.; Weigman, V.J.; Fan, C.; Sawyer, L.R.; He, X.; Troester, M.A.; Sartor, C.I.; Rieger-House, T.; Bernard, P.S.; Carey, L.A.; et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genom. 2007, 8, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anido, J.; Matar, P.; Albanell, J.; Guzman, M.; Rojo, F.; Arribas, J.; Averbuch, S.; Baselga, J. ZD1839, a specific epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, induces the formation of inactive EGFR/HER2 and EGFR/HER3 heterodimers and prevents heregulin signaling in HER2-overexpressing breast cancer cells. Clin. Cancer Res. 2003, 9, 1274–1283. [Google Scholar] [PubMed]
- Bjorkelund, H.; Gedda, L.; Barta, P.; Malmqvist, M.; Andersson, K. Gefitinib induces epidermal growth factor receptor dimers which alters the interaction characteristics with (1)(2)(5)I-EGF. PLoS ONE 2011, 6, e24739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dua, R.; Zhang, J.; Nhonthachit, P.; Penuel, E.; Petropoulos, C.; Parry, G. EGFR over-expression and activation in high HER2, ER negative breast cancer cell line induces trastuzumab resistance. Breast Cancer Res. Treat. 2010, 122, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Brockhoff, G. Target HER four in breast cancer? Oncotarget 2019, 10, 3147–3150. [Google Scholar] [CrossRef] [Green Version]
- Witton, C.J.; Reeves, J.R.; Going, J.J.; Cooke, T.G.; Bartlett, J.M. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J. Pathol. 2003, 200, 290–297. [Google Scholar] [CrossRef]
- Carlsson, J.; Wester, K.; De La Torre, M.; Malmstrom, P.U.; Gardmark, T. EGFR-expression in primary urinary bladder cancer and corresponding metastases and the relation to HER2-expression. On the possibility to target these receptors with radionuclides. Radiol. Oncol. 2015, 49, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Day, K.C.; Lorenzatti Hiles, G.; Kozminsky, M.; Dawsey, S.J.; Paul, A.; Broses, L.J.; Shah, R.; Kunja, L.P.; Hall, C.; Palanisamy, N.; et al. HER2 and EGFR Overexpression Support Metastatic Progression of Prostate Cancer to Bone. Cancer Res. 2017, 77, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Yeo, S.K.; Guan, J.L. Breast Cancer: Multiple Subtypes within a Tumor? Trends Cancer 2017, 3, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Wicinski, J.; Cervera, N.; Finetti, P.; Hur, M.H.; Diebel, M.E.; Monville, F.; Dutcher, J.; et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009, 69, 1302–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duru, N.; Candas, D.; Jiang, G.; Li, J.J. Breast cancer adaptive resistance: HER2 and cancer stem cell repopulation in a heterogeneous tumor society. J. Cancer Res. Clin. Oncol. 2014, 140, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Karnoub, A.E.; Dash, A.B.; Vo, A.P.; Sullivan, A.; Brooks, M.W.; Bell, G.W.; Richardson, A.L.; Polyak, K.; Tubo, R.; Weinberg, R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007, 449, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Wicha, M.S. HER2 and breast cancer stem cells: More than meets the eye. Cancer Res. 2013, 73, 3489–3493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Huang, W.; Tan, Y.; Jin, X.; Dua, R.; Penuel, E.; Mukherjee, A.; Sperinde, J.; Pannu, H.; Chenna, A.; et al. A novel proximity assay for the detection of proteins and protein complexes: Quantitation of HER1 and HER2 total protein expression and homodimerization in formalin-fixed, paraffin-embedded cell lines and breast cancer tissue. Diagn. Mol. Pathol. 2009, 18, 11–21. [Google Scholar] [CrossRef]
- Yang, T.; Xu, L.; Liu, S.; Shen, Y.; Huang, L.; Zhang, L.; Ding, S.; Cheng, W. Amplified fluorescence imaging of HER2 dimerization on cancer cells by using a co-localization triggered DNA nanoassembly. Mikrochim. Acta 2019, 186, 439. [Google Scholar] [CrossRef]
- Doh, J.K.; White, J.D.; Zane, H.K.; Chang, Y.H.; Lopez, C.S.; Enns, C.A.; Beatty, K.E. VIPER is a genetically encoded peptide tag for fluorescence and electron microscopy. Proc. Natl. Acad. Sci. USA 2018, 115, 12961–12966. [Google Scholar] [CrossRef] [Green Version]
- Weitsman, G.; Barber, P.R.; Nguyen, L.K.; Lawler, K.; Patel, G.; Woodman, N.; Kelleher, M.T.; Pinder, S.E.; Rowley, M.; Ellis, P.A.; et al. HER2-HER3 dimer quantification by FLIM-FRET predicts breast cancer metastatic relapse independently of HER2 IHC status. Oncotarget 2016, 7, 51012–51026. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.; Kim, W.; Kim, L.K.; VanHouten, J.; Wysolmerski, J.J. HER2 signaling regulates HER2 localization and membrane retention. PLoS ONE 2017, 12, e0174849. [Google Scholar] [CrossRef] [Green Version]
- Byrne, P.O.; Hristova, K.; Leahy, D.J. EGFR forms ligand-independent oligomers that are distinct from the active state. J. Biol. Chem. 2020, 295, 13353–13362. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, S.; Kim, D.K.; Jeong, M.G.; Noh, J.; Kwon, Y.; Zhou, K.; Lee, N.K.; Ryu, S.H. Direct visualization of single-molecule membrane protein interactions in living cells. PLoS Biol. 2018, 16, e2006660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peckys, D.B.; Korf, U.; de Jonge, N. Local variations of HER2 dimerization in breast cancer cells discovered by correlative fluorescence and liquid electron microscopy. Sci. Adv. 2015, 1, e1500165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsemarz, A.; Lasko, P.; Fagotto, F. Limited significance of the in situ proximity ligation assay. bioRxiv 2018. [Google Scholar] [CrossRef]
- Stanly, T.A.; Fritzsche, M.; Banerji, S.; Garcia, E.; Bernardino de la Serna, J.; Jackson, D.G.; Eggeling, C. Critical importance of appropriate fixation conditions for faithful imaging of receptor microclusters. Biol. Open 2016, 5, 1343–1350. [Google Scholar] [CrossRef] [Green Version]
- Chung, I.; Reichelt, M.; Shao, L.; Akita, R.W.; Koeppen, H.; Rangell, L.; Schaefer, G.; Mellman, I.; Sliwkowski, M.X. High cell-surface density of HER2 deforms cell membranes. Nat. Commun. 2016, 7, 12742. [Google Scholar] [CrossRef] [Green Version]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 2004, 37, 790–796. [Google Scholar] [CrossRef]
- Giepmans, B.N.; Deerinck, T.J.; Smarr, B.L.; Jones, Y.Z.; Ellisman, M.H. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods 2005, 2, 743–749. [Google Scholar] [CrossRef]
- Deerinck, T.J. The Application of Fluorescent Quantum Dots to Confocal, Multiphoton, and Electron Microscopic Imaging. Toxicol. Pathol. 2008, 36, 112–116. [Google Scholar] [CrossRef] [Green Version]
- Alansary, D.; Peckys, D.B.; Niemeyer, B.A.; de Jonge, N. Detecting single ORAI1 proteins within the plasma membrane reveals higher-order channel complexes. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Blach, P.; Keskin, S.; de Jonge, N. Graphene Enclosure of Chemically Fixed Mammalian Cells for Liquid-Phase Electron Microscopy. J. Vis. Exp. 2020, 163, e61458-61451-61420. [Google Scholar] [CrossRef] [PubMed]
- Dahmke, I.N.; Verch, A.; Hermannsdorfer, J.; Peckys, D.B.; Weatherup, R.S.; Hofmann, S.; de Jonge, N. Graphene Liquid Enclosure for Single-Molecule Analysis of Membrane Proteins in Whole Cells Using Electron Microscopy. ACS Nano 2017, 11, 11108–11117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagnato, P.; Castagnino, A.; Cortese, K.; Bono, M.; Grasso, S.; Bellese, G.; Daniele, T.; Lundmark, R.; Defilippi, P.; Castagnola, P.; et al. Cooperative but distinct early co-signaling events originate from ERBB2 and ERBB1 receptors upon trastuzumab treatment in breast cancer cells. Oncotarget 2017, 8, 60109–60122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peckys, D.B.; Quint, C.; Jonge, N. Determining the Efficiency of Single Molecule Quantum Dot Labeling of HER2 in Breast Cancer Cells. Nano Lett. 2020, 20, 7948–7955. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.M.; Albrecht, D.; Culley, S.; Jacobs, C.; Marsh, M.; Mercer, J.; Henriques, R. Fix Your Membrane Receptor Imaging: Actin Cytoskeleton and CD4 Membrane Organization Disruption by Chemical Fixation. Front. Immunol. 2019, 10, 675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whelan, D.R.; Bell, T.D. Image artifacts in single molecule localization microscopy: Why optimization of sample preparation protocols matters. Sci. Rep. 2015, 5, 7924. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.A.; Suzuki, K.G.; Shirai, Y.M.; Shibutani, S.T.; Miyahara, M.S.; Tsuboi, H.; Yahara, M.; Yoshimura, A.; Mayor, S.; Fujiwara, T.K.; et al. Membrane molecules mobile even after chemical fixation. Nat. Methods 2010, 7, 865–866. [Google Scholar] [CrossRef]
- Tagliaferro, P.; Tandler, C.J.; Ramos, A.J.; Pecci Saavedra, J.; Brusco, A. Immunofluorescence and glutaraldehyde fixation. A new procedure based on the Schiff-quenching method. J. Neurosci. Methods 1997, 77, 191–197. [Google Scholar] [CrossRef]
- Arnspang, E.C.; Brewer, J.R.; Lagerholm, B.C. Multi-Color Single Particle Tracking with Quantum Dots. PLoS ONE 2012, 7, e48521. [Google Scholar] [CrossRef]
- Byers, R.J.; Hitchman, E.R. Quantum Dots Brighten Biological Imaging. Prog. Histochem. Cyto. 2011, 45, 201–237. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Chaudry, Q.; Shen, C.; Kong, K.Y.; Zhau, H.E.; Chung, L.W.; Petros, J.A.; O’Regan, R.M.; Yezhelyev, M.V.; Simons, J.W.; et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat. Protoc. 2007, 2, 1152–1165. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.Q.; Lam, W.Y.; Hatch, E.W.; Lidke, D.S. Quantum dots for quantitative imaging: From single molecules to tissue. Cell Tissue Res. 2015, 360, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Sosinsky, G.E.; Giepmans, B.N.G.; Deerinck, T.J.; Gaietta, G.M.; Ellisman, M.H. Markers for correlated light and electron microscopy. In Cellular Electron Microscopy; Elsevier Academic Press Inc: San Diego, CA, USA, 2007; Volume 79, pp. 575–591. [Google Scholar]
- Weinberg, F.; Han, M.K.L.; Dahmke, I.N.; Del Campo, A.; de Jonge, N. Anti-correlation of HER2 and focal adhesion complexes in the plasma membrane. PLoS ONE 2020, 15, e0234430. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; VanHouten, J.N.; Kim, W.; Dann, P.; Sullivan, C.; Choi, J.; Sneddon, W.B.; Friedman, P.A.; Wysolmerski, J.J. The scaffolding protein NHERF1 regulates the stability and activity of the tyrosine kinase HER2. J. Biol. Chem. 2017, 292, 6555–6568. [Google Scholar] [CrossRef] [Green Version]
- Bertelsen, V.; Stang, E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes 2014, 4, 424–446. [Google Scholar] [CrossRef] [Green Version]
- Nagy, P.; Vereb, G.; Sebestyen, Z.; Horvath, G.; Lockett, S.J.; Damjanovich, S.; Park, J.W.; Jovin, T.M.; Szollosi, J. Lipid rafts and the local density of ErbB proteins influence the biological role of homo- and heteroassociations of ErbB2. J. Cell Sci. 2002, 115, 4251–4262. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.B.; Berger, C.; Rodland, M.S.; Hasmann, M.; Stang, E.; Madshus, I.H. Pertuzumab increases epidermal growth factor receptor down-regulation by counteracting epidermal growth factor receptor-ErbB2 heterodimerization. Mol. Cancer 2009, 8, 1885–1892. [Google Scholar] [CrossRef] [Green Version]
- Nami, B.; Wang, Z.X. HER2 in Breast Cancer Stemness: A Negative Feedback Loop towards Trastuzumab Resistance. Cancers 2017, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Peckys, D.B.; Korf, U.; Wiemann, S.; de Jonge, N. Liquid-phase electron microscopy of molecular drug response in breast cancer cells reveals irresponsive cell subpopulations related to lack of HER2 homodimers. Mol. Biol. Cell 2017, 28, 3193–3202. [Google Scholar] [CrossRef]
- Lu, Y.; Zi, X.; Zhao, Y.; Mascarenhas, D.; Pollak, M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J. Natl. Cancer Inst. 2001, 93, 1852–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shattuck, D.L.; Miller, J.K.; Carraway, K.L., 3rd; Sweeney, C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008, 68, 1471–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, S.P.; Han, J.Z.R.; Portman, N.; Nobis, M.; Hastings, J.F.; Murphy, K.J.; Latham, S.L.; Cadell, A.L.; Miladinovic, D.; Marriott, G.R.; et al. Targeting promiscuous heterodimerization overcomes innate resistance to ERBB2 dimerization inhibitors in breast cancer. Breast Cancer Res. 2019, 21, 43. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, H.J.; Poulsen, T.T.; Dahlman, A.; Kjaer, I.; Koefoed, K.; Sen, J.W.; Weilguny, D.; Bjerregaard, B.; Andersen, C.R.; Horak, I.D.; et al. Pan-HER, an Antibody Mixture Simultaneously Targeting EGFR, HER2, and HER3, Effectively Overcomes Tumor Heterogeneity and Plasticity. Clin. Cancer Res. 2015, 21, 4110–4122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saura, C.; Oliveira, M.; Feng, Y.-H.; Dai, M.-S.; Hurvitz, S.A.; Kim, S.-B.; Moy, B.; Delaloge, S.; Gradishar, W.J.; Masuda, N.; et al. Neratinib + capecitabine versus lapatinib + capecitabine in patients with HER2+ metastatic breast cancer previously treated with ≥ 2 HER2-directed regimens: Findings from the multinational, randomized, phase III NALA trial. J. Clin. Oncol. 2019, 37, 1002. [Google Scholar] [CrossRef]
- Llombart-Cussac, A.; Cortés, J.; Paré, L.; Galván, P.; Bermejo, B.; Martínez, N.; Vidal, M.; Pernas, S.; López, R.; Muñoz, M.; et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): An open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017, 18, 545–554. [Google Scholar] [CrossRef]
- Ma, F.; Ouyang, Q.; Li, W.; Jiang, Z.; Tong, Z.; Liu, Y.; Li, H.; Yu, S.; Feng, J.; Wang, S.; et al. Pyrotinib or Lapatinib Combined With Capecitabine in HER2–Positive Metastatic Breast Cancer With Prior Taxanes, Anthracyclines, and/or Trastuzumab: A Randomized, Phase II Study. J. Clin. Oncol. 2019, 37, 2610–2619. [Google Scholar] [CrossRef]
- Patel, A.; Unni, N.; Peng, Y. The Changing Paradigm for the Treatment of HER2-Positive Breast Cancer. Cancers 2020, 12, 81. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [Green Version]
- Gianni, L.; Pienkowski, T.; Im, Y.H.; Tseng, L.M.; Liu, M.C.; Lluch, A.; Staroslawska, E.; de la Haba-Rodriguez, J.; Im, S.A.; Pedrini, J.L.; et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): A multicentre, open-label, phase 2 randomised trial. Lancet Oncol. 2016, 17, 791–800. [Google Scholar] [CrossRef]
- Peckys, D.B.; Hirsch, D.; Gaiser, T.; de Jonge, N. Visualisation of HER2 homodimers in single cells from HER2 overexpressing primary formalin fixed paraffin embedded tumour tissue. Mol. Med. 2019, 25, 42. [Google Scholar] [CrossRef] [PubMed]
- Peckys, D.B.; de Jonge, N. Studying the Stoichiometry of Epidermal Growth Factor Receptor in Intact Cells using Correlative Microscopy. J. Vis. Exp. 2015, e53186. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Cell Subtype | Cells | Images | Imaged Area [µm2] | Particles QD565 | HER2-QD565density/µm2 (Mean ± STD) × 102 (QDs/µm2) | Particles QD655 | EGFR-QD655density/µm2 Mean ± STD × 101 (QDs/µm2) |
|---|---|---|---|---|---|---|---|
| Bulk, clustered regions | 31 | 68 | 196 | 52,744 | 2.7 ± 1.2 | 1301 | 0.6 ± 0.6 |
| Bulk, large membrane protrusions | 21 | 45 | 128 | 128,847 | 9.9 ± 4.5 | 2478 | 2 ± 1 |
| Bulk, flat regions | 15 | 31 | 89 | 18,212 | 2.0 ± 0.7 | 448 | 0.5 ± 0.3 |
| EGFR enriched, large membrane protrusions | 10 | 26 | 72 | 52,675 | 7.0 ± 2.3 | 3606 | 5 ± 2 |
| EGFR enriched, clustered regions | 10 | 34 | 98 | 26,047 | 2.7 ± 1.2 | 1342 | 1.3 ± 0.7 |
| Background (substrate) | n.a. | 15 | 43 | 2531 | 0.6 ± 0.2 | 102 | 0.2 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weinberg, F.; Peckys, D.B.; de Jonge, N. EGFR Expression in HER2-Driven Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 9008. https://doi.org/10.3390/ijms21239008
Weinberg F, Peckys DB, de Jonge N. EGFR Expression in HER2-Driven Breast Cancer Cells. International Journal of Molecular Sciences. 2020; 21(23):9008. https://doi.org/10.3390/ijms21239008
Chicago/Turabian StyleWeinberg, Florian, Diana B. Peckys, and Niels de Jonge. 2020. "EGFR Expression in HER2-Driven Breast Cancer Cells" International Journal of Molecular Sciences 21, no. 23: 9008. https://doi.org/10.3390/ijms21239008