Abstract
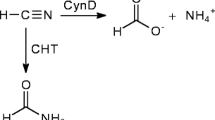
Recombinant forms of three cyanide-degrading nitrilases, CynD from Bacillus pumilus C1, CynD from Pseudomonas stutzeri, and CHT from Gloeocercospora sorghi, were prepared after their genes were cloned with C-terminal hexahistidine purification tags and expressed in Escherichia coli, and the enzymes purified using nickel-chelate affinity chromatography. The enzymes were compared with respect to their pH stability, thermostability, metal tolerance, and kinetic constants. The two bacterial genes, both cyanide dihydratases, were similar with respect to pH range, retaining greater than 50% activity between pH 5.2 and pH 8 and kinetic properties, having similar Km (6–7 mM) and Vmax (0.1 mmol min−1 mg−1). They also exhibited similar metal tolerances. However, the fungal CHT enzyme had notably higher Km (90 mM) and Vmax (4 mmol min−1 mg−1) values. Its pH range was slightly more alkaline (retaining nearly full activity above 8.5), but exhibited a lower thermal tolerance. CHT was less sensitive to Hg2+ and more sensitive to Pb2+ than the CynD enzymes. These data describe, in part, the current limits that exist for using nitrilases as agents in the bioremediation of cyanide-containing waste effluent, and may help serve to determine where and under what conditions these nitrilases may be applied.





Similar content being viewed by others
References
Basheer S, Kut ÖM, Prenosil JE, Bourne JR (1993) Development of an enzyme membrane reactor for treatment of cyanide-containing wastewaters from the food industry. Biotechnol Bioeng 41:465–473
Brenner C (2002) Catalysis in the nitrilase superfamily. Curr Opin Struct Biol 12:775–782
Brown DT, Turner PD, O’Reilly C (1995) Expression of the cyanide hydratase enzyme from Fusarium lateritium in Escherichia coli and identification of an essential cysteine residue. FEMS Microbiol Lett 134:143–146
Cluness MM, Turner PD, Clements E, Brown DT, O’Reilly C (1993) Purification and properties of cyanide hydratase from Fusarium lateritium and analysis of corresponding chyI gene. J Gen Microbiol 139:1807–1815
Dumestre A, Chone T, Portal JM, Gerard M, Berthelin J (1997) Cyanide degradation under alkaline conditions by a strain of Fusarium solani from contaminated soils. Appl Environ Microbiol 63:2729–2734
Fisher FB, Brown JS (1952) Colorimetric determination of cyanide in stack gas and waste water. Anal Chem 24:1440–1444
Fry WE, Munch DC (1975) Hydrogen cyanide detoxification by Gloeocercospora sorghi. Physiol Plant Pathol 7:23–33
Goldlust A, Bohak Z (1989) Induction, purification, and characterization of the nitrilase of Fusarium oxysporum f. sp. melonis. Biotechnol Appl Biochem 11:581–601
Harper DB (1977) Microbial metabolism of aromatic nitriles. Enzymology of C–N cleavage by Nocardia sp. (Rhodochrous group) N.C.I.B. 11216. Biochem J 165:309–319
Hook RH, Robinson WG (1964) Ricinine nitrilase II purification and properties. J Biol Chem 239:4263–4267
Ingvorsen K, Højer-Pedersen B, Godtrfedsen SE (1991) Novel cyanide-hydrolyzing enzyme from Alcaligenes xylosoxidans subsp denitrificans. Appl Environ Microbiol 57:1783–1789
Ingvorsen K, Godtfredsen SE, Højer-Pedersen B, Novo Nordisk (1992) Microbial cyanide converting enzymes, their production and use. United States Patent 5,116,744
Jandhyala DM, Berman M, Meyers PR, Sewell T, Willson RC, Benedik MJ (2003) CynD, the cyanide dihydratase from Bacillus pumilus: gene cloning and structural studies. Appl Environ Microbiol 69:4794–4805
Kobayashi M, Yanaka N, Nagasawa T, Yamada H (1992) Primary structure of an aliphatic nitrile-degrading enzyme, aliphatic nitrilase, from Rhodococcus rhodochrous K22 and expression of its gene and identification of its active site residue. Biochem 31:9000–9007
Kobayashi M, Goda M, Sakayu S (1998) Nitrilase catalyzes amide hydrolysis as well as nitrile hydrolysis. Biochem Biophys Res Commun 253:662–666
Meyers PR, Rawlings DE, Woods DR, GG Lindsey (1993) Isolation and characterization of a cyanide dihydratase for Bacillus pumilus C1. J Bacteriol 175:6105–6112
Mudder T, Whitlock J (1984) Biological treatment of cyanidation wastewaters. Miner Metall Process 1:161–164
Nagasawa T, Mauger J, Yamada H (1990) A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3. Purification and characterization. Eur J Biochem 194:765–772
Nakai T, Hasgawa T, Yamashita E, Yamamoto M, Kamasuka T, Ueki T, Nanba H, Ikenaka Y, Takahashi S, Sato M, Tsukihara T (2000) Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 8:729–737
Nolan LM, Harnedy PA, Turner P, Hearne AB, O’Reilly C (2003) The cyanide hydratase enzyme of Fusarium lateritium also has nitrilase activity. FEMS Microbiol Lett 221:161–165
O’Reilly C, Turner PD (2003) The nitrilase family of CN hydrolyzing enzymes—a comparative study. J Appl Microbiol 95:1161–1174
Richardson K, Clarke P, Imperial Chemical Industries (1993) Production of cyanide hydratase. United States Patent 5, 219, 750
Sewell BT, Meyers P, Berman M, Jandhyala DM, Benedik MJ (2003) The cyanide degrading nitrilase from Pseudomonas stutzeri AK61 is a twofold symmetric, 14-subunit spiral. Structure 11:1413–1422
Sexton AC, Howlett BJ (2000) Characterisation of a cyanide hydratase gene in the phytopathogenic fungus Leptosphaeria maculans. Mol Gen Genet 263:463–470
Stevenson DE, Feng R, Dumas F, Groleau D, Mihoc A, Storer AC (1992) Mechanistic and structural studies on Rhodococcus ATCC 39484 nitrilase. Biotechnol Appl Biochem 15:283–302
Wang P, VanEtten HD (1992) Cloning an properties of a cyanide hydratase gene from the phytopathogenic fungus Gloeocercospora sorghi. Biochem Biophys Res Commun 187:1048–1054
Wang P, Matthews DE, VanEtten HD (1992) Purification and characterization of cyanide hydratase from the phytopathogenic fungus Gloeocercospora sorghi. Arch Biochem Biophys 298:569–575
Watanabe A, Yano K, Ikebukuro K, Karube I (1998a) Cyanide hydrolysis in a cyanide-degrading bacterium, Pseudomonas stutzeri AK61, by cyanidase. Microbiology 144:1677–1682
Watanabe A, Yano K, Ikebukuro K, Karube I (1998b) Cloning and expression of a gene encoding cyanidase from Pseudomonas stutzeri AK61. Appl Microbiol Biotechnol 50:93–97
Watanabe A, Yano K, Ikebukuro K, Karube I (1998c) Investigation of the potential active site of a cyanide dihydratase using site-directed mutagenesis. Biochim Biophys Acta 1382:1–4
Yanase H, Sakamoto A, Okamoto K, Kita K, Sato Y (2000) Degradation of the metal-cyano complex tetracyanonickelate (II) by Fusarium oxysporum N-10. Appl Microbiol Biotechnol 53:328–334
Acknowledgements
We gratefully acknowledge support from the Robert A. Welch Foundation, the University of Houston Environmental Institute, and the Gulf Coast Hazardous Substance Research Center, #069UHH0789, to M.J.B. and from the Wellcome Trust to B.T.S. for support of this project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jandhyala, D.M., Willson, R.C., Sewell, B.T. et al. Comparison of cyanide-degrading nitrilases. Appl Microbiol Biotechnol 68, 327–335 (2005). https://doi.org/10.1007/s00253-005-1903-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-1903-8