Abstract
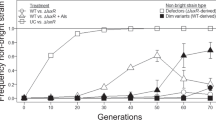
Pathogens often infect hosts through collective actions: they secrete growth-promoting compounds or virulence factors, or evoke host reactions that fuel the colonization of the host. Such behaviours are vulnerable to the rise of mutants that benefit from the collective action without contributing to it; how these behaviours can be evolutionarily stable is not well understood1. We address this question using the intestinal pathogen Salmonella enterica serovar Typhimurium (hereafter termed S. typhimurium), which manipulates its host to induce inflammation, and thereby outcompetes the commensal microbiota2,3. Notably, the virulence factors needed for host manipulation are expressed in a bistable fashion, leading to a slow-growing subpopulation that expresses virulence genes, and a fast-growing subpopulation that is phenotypically avirulent4,5. Here we show that the expression of the genetically identical but phenotypically avirulent subpopulation is essential for the evolutionary stability of virulence in this pathogen. Using a combination of mathematical modelling, experimental evolution and competition experiments we found that within-host evolution leads to the emergence of mutants that are genetically avirulent and fast-growing. These mutants are defectors that exploit inflammation without contributing to it. In infection experiments initiated with wild-type S. typhimurium, defectors increase only slowly in frequency. In a genetically modified S. typhimurium strain in which the phenotypically avirulent subpopulation is reduced in size, defectors rise more rapidly, inflammation ceases prematurely, and S. typhimurium is quickly cleared from the gut. Our results establish that host manipulation by S. typhimurium is a cooperative trait that is vulnerable to the rise of avirulent defectors; the expression of a phenotypically avirulent subpopulation that grows as fast as defectors slows down this process, and thereby promotes the evolutionary stability of virulence. This points to a key role of bistable virulence gene expression in stabilizing cooperative virulence and may lead the way to new approaches for controlling pathogens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
West, S. A., Griffin, A. S., Gardner, A. & Diggle, S. P. Social evolution theory for microorganisms. Nature Rev. Microbiol. 4, 597–607 (2006)
Stecher, B. et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, e244 (2007)
Kaiser, P., Diard, M., Stecher, B. & Hardt, W. D. The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen’s virulence factors, and the host’s mucosal immune response. Immunol. Rev. 245, 56–83 (2012)
Ackermann, M. et al. Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987–990 (2008)
Sturm, A. et al. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7, e1002143 (2011)
Casadevall, A. & Pirofski, L. A. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 67, 3703–3713 (1999)
van Baalen, M. & Sabelis, M. W. The dynamics of multiple infection and the evolution of virulence. Am. Nat. 146, 881–910 (1995)
Lysenko, E. S., Lijek, R. S., Brown, S. P. & Weiser, J. N. Within-host competition drives selection for the capsule virulence determinant of Streptococcus pneumoniae . Curr. Biol. 20, 1222–1226 (2010)
Brown, S. P. Cooperation and conflict in host-manipulating parasites. Proc. R. Soc. Lond. B 266, 1899–1904 (1999)
Brown, S. P., Hochberg, M. E. & Grenfell, B. T. Does multiple infection select for raised virulence? Trends Microbiol. 10, 401–405 (2002)
Buckling, A. & Brockhurst, M. A. Kin selection and the evolution of virulence. Heredity 100, 484–488 (2008)
Harrison, F., Browning, L. E., Vos, M. & Buckling, A. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 4, 21 (2006)
Köhler, T., Buckling, A. & van Delden, C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl Acad. Sci. USA 106, 6339–6344 (2009)
Rumbaugh, K. P. et al. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19, 341–345 (2009)
Raymond, B., West, S. A., Griffin, A. S. & Bonsall, M. B. The dynamics of cooperative bacterial virulence in the field. Science 337, 85–88 (2012)
Brown, S. P., Le Chat, L. & Taddei, F. Evolution of virulence: triggering host inflammation allows invading pathogens to exclude competitors. Ecol. Lett. 11, 44–51 (2008)
Endt, K. et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6, e1001097 (2010)
Suar, M. et al. Accelerated type III secretion system 2-dependent enteropathogenesis by a Salmonella enterica serovar enteritidis PT4/6 strain. Infect. Immun. 77, 3569–3577 (2009)
Golubeva, Y. A., Sadik, A. Y., Ellermeier, J. R. & Slauch, J. M. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190, 79–90 (2012)
Baxter, M. A., Fahlen, T. F., Wilson, R. L. & Jones, B. D. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71, 1295–1305 (2003)
Cremer, J., Melbinger, A. & Frey, E. Growth dynamics and the evolution of cooperation in microbial populations. Sci. Rep. 2, 281 (2012)
Lawley, T. D. et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 76, 403–416 (2008)
Bush, K. et al. Tackling antibiotic resistance. Nature Rev. Microbiol. 9, 894–896 (2011)
R Devlopment Core Team. A Language and Environment for Statistical Computing http://www.R-project.org (R Foundation for Statistical Computing, 2010)
Soetaert, K., Petzoldt, T. & Setzer, R. W. Solving differential equations in R: Package deSolve. J. Stat. Softw. 33, 1–25 (2010)
Hoiseth, S. K. & Stocker, B. A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981)
Kaniga, K., Bossio, J. C. & Galan, J. E. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13, 555–568 (1994)
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000)
Santiviago, C. A. et al. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5, e1000477 (2009)
Browne, S. H., Hasegawa, P., Okamoto, S., Fierer, J. & Guiney, D. G. Identification of Salmonella SPI-2 secretion system components required for SpvB-mediated cytotoxicity in macrophages and virulence in mice. FEMS Immunol. Med. Microbiol. 52, 194–201 (2008)
Hensel, M. et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30, 163–174 (1998)
Suar, M. et al. Accelerated type III secretion system 2-dependent enteropathogenesis by a Salmonella enterica serovar enteritidis PT4/6 strain. Infect. Immun. 77, 3569–3577 (2009)
Schechter, L. M. & Lee, C. A. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40, 1289–1299 (2001)
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995)
Sturm, A. et al. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7, e1002143 (2011)
Stecher, B. et al. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72, 4138–4150 (2004)
Hautefort, I., Proenca, M. J. & Hinton, J. C. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69, 7480–7491 (2003)
Gil, D. & Bouche, J. P. ColE1-type vectors with fully repressible replication. Gene 105, 17–22 (1991)
Endt, K. et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhoea. PLoS Pathog. 6, e1001097 (2010)
Nordentoft, S., Christensen, H. & Wegener, H. C. Evaluation of a fluorescence-labelled oligonucleotide probe targeting 23S rRNA for in situ detection of Salmonella serovars in paraffin-embedded tissue sections and their rapid identification in bacterial smears. J. Clin. Microbiol. 35, 2642–2648 (1997)
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012)
Barthel, M. et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858 (2003)
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009)
Acknowledgements
We thank E. Slack, R. Kümmerli, M. Sellin and L. Robert for comments on the manuscript, G. Paul for input on the modelling, and Hardt laboratory members for discussions. M.D. was supported in part by the Fondation pour la Recherche Médicale. M.A., R.R.R. and W.-D.H. were supported by grants from the Swiss National Science Foundation.
Author information
Authors and Affiliations
Contributions
M.D., V.G. and R.R.R. conceived and analysed the mathematical simulations. M.D., V.G. and R.R.R. wrote the theoretical part of the paper. M.D., W.-D.H., L.M. (Supplementary Figs 14 and 20), M.N.P.R.-E (Supplementary Fig. 8) and M.A. designed the experiments and analysed the data. M.D., W.-D.H. and M.A. wrote the paper. M.D. and L.M. (Supplementary Figs 14 and 20) performed the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
This file contains Supplementary Text and Data, Supplementary Table 1, Supplementary Figures 1-20 and Supplementary References. (PDF 1368 kb)
Rights and permissions
About this article
Cite this article
Diard, M., Garcia, V., Maier, L. et al. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494, 353–356 (2013). https://doi.org/10.1038/nature11913
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11913
This article is cited by
-
Transcription-driven DNA supercoiling counteracts H-NS-mediated gene silencing in bacterial chromatin
Nature Communications (2024)
-
A genetic switch controls Pseudomonas aeruginosa surface colonization
Nature Microbiology (2023)
-
Synthetic genetic oscillators demonstrate the functional importance of phenotypic variation in pneumococcal-host interactions
Nature Communications (2023)
-
Dynamic altruistic cooperation within breast tumors
Molecular Cancer (2023)
-
Impact of horizontal gene transfer on emergence and stability of cooperative virulence in Salmonella Typhimurium
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.