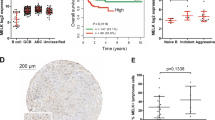
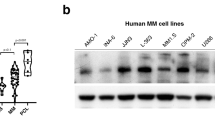
Abstract
We searched for genetic alterations in human B cell lymphoma that affect the ubiquitin-proteasome system. This approach identified FBXO25 within a minimal common region of frequent deletion in mantle cell lymphoma (MCL). FBXO25 encodes an orphan F-box protein that determines the substrate specificity of the SCF (SKP1–CUL1–F-box)FBXO25 ubiquitin ligase complex. An unbiased screen uncovered the prosurvival protein HCLS1-associated protein X-1 (HAX-1) as the bona fide substrate of FBXO25 that is targeted after apoptotic stresses. Protein kinase Cδ (PRKCD) initiates this process by phosphorylating FBXO25 and HAX-1, thereby spatially directing nuclear FBXO25 to mitochondrial HAX-1. Our analyses in primary human MCL identify monoallelic loss of FBXO25 and stabilizing HAX1 phosphodegron mutations. Accordingly, FBXO25 re-expression in FBXO25-deleted MCL cells promotes cell death, whereas expression of the HAX-1 phosphodegron mutant inhibits apoptosis. In addition, knockdown of FBXO25 significantly accelerated lymphoma development in Eμ-Myc mice and in a human MCL xenotransplant model. Together we identify a PRKCD-dependent proapoptotic mechanism controlling HAX-1 stability, and we propose that FBXO25 functions as a haploinsufficient tumor suppressor and that HAX1 is a proto-oncogene in MCL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Shaffer, A.L. III., Young, R.M. & Staudt, L.M. Pathogenesis of human B cell lymphomas. Annu. Rev. Immunol. 30, 565–610 (2012).
Puebla-Osorio, N. & Zhu, C. DNA damage and repair during lymphoid development: antigen receptor diversity, genomic integrity and lymphomagenesis. Immunol. Res. 41, 103–122 (2008).
Suzuki, Y. et al. HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J. Immunol. 158, 2736–2744 (1997).
Yap, S.V., Koontz, J.M. & Kontrogianni-Konstantopoulos, A. HAX-1: a family of apoptotic regulators in health and disease. J. Cell. Physiol. 226, 2752–2761 (2011).
Chao, J.R. et al. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 452, 98–102 (2008).
Klein, C. Genetic defects in severe congenital neutropenia: emerging insights into life and death of human neutrophil granulocytes. Annu. Rev. Immunol. 29, 399–413 (2011).
Peckl-Schmid, D. et al. HAX1 deficiency: impact on lymphopoiesis and B-cell development. Eur. J. Immunol. 40, 3161–3172 (2010).
Kwiecinska, A. et al. HAX-1 expression in human B lymphoma. Leukemia 25, 868–872 (2011).
Wei, X.J. et al. Expression of HAX-1 in human colorectal cancer and its clinical significance. Tumour Biol. 35, 1411–1415 (2014).
Li, M. et al. Analysis of HAX-1 gene expression in esophageal squamous cell carcinoma. Diagn. Pathol. 8, 47 (2013).
Han, J. et al. Deregulation of mitochondrial membrane potential by mitochondrial insertion of granzyme B and direct Hax-1 cleavage. J. Biol. Chem. 285, 22461–22472 (2010).
Li, B. et al. Hax-1 is rapidly degraded by the proteasome dependent on its PEST sequence. BMC Cell Biol. 13, 20 (2012).
Griner, E.M. & Kazanietz, M.G. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer 7, 281–294 (2007).
Yoshida, K. PKCdelta signaling: mechanisms of DNA damage response and apoptosis. Cell. Signal. 19, 892–901 (2007).
Miyamoto, A. et al. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature 416, 865–869 (2002).
DeVries-Seimon, T.A., Ohm, A.M., Humphries, M.J. & Reyland, M.E. Induction of apoptosis is driven by nuclear retention of protein kinase Cδ. J. Biol. Chem. 282, 22307–22314 (2007).
Petroski, M.D. & Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005).
Skaar, J.R., D'Angiolella, V., Pagan, J.K. & Pagano, M. SnapShot: F box proteins II. Cell 137, 1358, 1358.e1 (2009).
Bassermann, F., Eichner, R. & Pagano, M. The ubiquitin proteasome system—implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta 1843, 150–162 (2014).
Skaar, J.R., Pagan, J.K. & Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013).
Rubio-Moscardo, F. et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood 105, 4445–4454 (2005).
Mestre-Escorihuela, C. et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood 109, 271–280 (2007).
Salaverria, I. et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression–based proliferation signature. J. Clin. Oncol. 25, 1216–1222 (2007).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 (2011).
Cheung, K.J. et al. Genome-wide profiling of follicular lymphoma by array comparative genomic hybridization reveals prognostically significant DNA copy number imbalances. Blood 113, 137–148 (2009).
Toujani, S. et al. High resolution genome-wide analysis of chromosomal alterations in Burkitt's lymphoma. PLoS ONE 4, e7089 (2009).
Dreyling, M. et al. Update on the molecular pathogenesis and clinical treatment of mantle cell lymphoma: report of the 10th annual conference of the European Mantle Cell Lymphoma Network. Leuk. Lymphoma 52, 2226–2236 (2011).
Schraders, M. et al. Novel chromosomal imbalances in mantle cell lymphoma detected by genome-wide array-based comparative genomic hybridization. Blood 105, 1686–1693 (2005).
Flordal Thelander, E. et al. Detailed assessment of copy number alterations revealing homozygous deletions in 1p and 13q in mantle cell lymphoma. Leuk. Res. 31, 1219–1230 (2007).
Tagawa, H. et al. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene 24, 1348–1358 (2005).
Martinez-Climent, J.A. et al. Loss of a novel tumor suppressor gene locus at chromosome 8p is associated with leukemic mantle cell lymphoma. Blood 98, 3479–3482 (2001).
Hagens, O., Minina, E., Schweiger, S., Ropers, H.H. & Kalscheuer, V. Characterization of FBX25, encoding a novel brain-expressed F-box protein. Biochim. Biophys. Acta 1760, 110–118 (2006).
Klein, C. et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease). Nat. Genet. 39, 86–92 (2007).
Fernández-Sáiz, V. et al. SCF-Fbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat. Cell Biol. 15, 72–81 (2013).
Bassermann, F. et al. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage–response checkpoint. Cell 134, 256–267 (2008).
Adams, J.M. et al. The c-Myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985).
Eischen, C.M., Weber, J.D., Roussel, M.F., Sherr, C.J. & Cleveland, J.L. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13, 2658–2669 (1999).
Vaux, D.L., Cory, S. & Adams, J.M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-Myc to immortalize pre-B cells. Nature 335, 440–442 (1988).
Hatzi, K. & Melnick, A. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 20, 343–352 (2014).
Souers, A.J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
Bassermann, F. et al. NIPA defines an SCF-type mammalian E3 ligase that regulates mitotic entry. Cell 122, 45–57 (2005).
Busino, L. et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904 (2007).
Bassermann, F. et al. Multisite phosphorylation of nuclear interaction partner of ALK (NIPA) at G2/M involves cyclin B1/Cdk1. J. Biol. Chem. 282, 15965–15972 (2007).
Volkmann, J. et al. High expression of crystallin αB represents an independent molecular marker for unfavourable ovarian cancer patient outcome and impairs TRAIL- and cisplatin-induced apoptosis in human ovarian cancer cells. Int. J. Cancer 132, 2820–2832 (2013).
Kraus, J.A., Dabbs, D.J., Beriwal, S. & Bhargava, R. Semi-quantitative immunohistochemical assay versus oncotype DX((R)) qRT-PCR assay for estrogen and progesterone receptors: an independent quality assurance study. Mod. Pathol. 25, 869–876 (2012).
Budczies, J. et al. Cutoff Finder: a comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS ONE 7, e51862 (2012).
Becker, K.F. et al. Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. J. Pathol. 211, 370–378 (2007).
Berg, D., Malinowsky, K., Reischauer, B., Wolff, C. & Becker, K.F. Use of formalin-fixed and paraffin-embedded tissues for diagnosis and therapy in routine clinical settings. Methods Mol. Biol. 785, 109–122 (2011).
Scuoppo, C. et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature 487, 244–248 (2012).
den Hollander, J. et al. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood 116, 1498–1505 (2010).
Acknowledgements
We thank J. Ihle (St. Jude Children's Research Hospital), V. Kalscheuer (Max Planck Institute for Molecular Genetics), M. Pagano (New York University School of Medicine) and A. Villunger (Medical University of Innsbruck) for reagents and cell lines; J. Martinez-Climent, M. Schraders, H. Tagawa, H. Kohlhammer and E. Flordal Thelander for sending the raw data of aCGH studies; G. Keller and K. Malinowski for suggestions; K.-F. Becker and C. Schott for help with extract preparation from MCL tissue samples; and R. Oellinger for help with the MCL tissue culture models. This work was supported by grants from the German Research Foundation (KE 222/-1 and SFB824 to U.K.; and BA 2851/4-1 and Emmy Noether Program grant BA 2851/3-1 to F.B.) and the German Cancer Aid (#109543 to F.B.).
Author information
Authors and Affiliations
Contributions
U.B., V.F.-S. and F.B. conceived and designed the research. U.B. and V.F.-S. performed most of the experiments with crucial help from A.-M.K., B.-S.T., V.T. and K.E. I.J., J.S. and Z.L. helped with the MCL xenotransplant model. M.R., A.R., W.K. and M.D. provided lymphoma samples. M.R. and A.R. performed IHC analyses. R.R. performed analyses of aCGH data. S.L. and B.K. performed mass spectrometry. C.M., A.-L.I., G.L., M.B., S.P. and M.L. provided critical reagents. U.B., V.F.-S., B.-S.T., K.E., S.L., I.J., C.P., U.K., B.K. and F.B. analyzed results. F.B. coordinated this work and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 (PDF 5360 kb)
Rights and permissions
About this article
Cite this article
Baumann, U., Fernández-Sáiz, V., Rudelius, M. et al. Disruption of the PRKCD–FBXO25–HAX-1 axis attenuates the apoptotic response and drives lymphomagenesis. Nat Med 20, 1401–1409 (2014). https://doi.org/10.1038/nm.3740
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3740
This article is cited by
-
Linking post-translational modifications and protein turnover by site-resolved protein turnover profiling
Nature Communications (2022)
-
Molecular determinants of outcomes in relapsed or refractory mantle cell lymphoma treated with ibrutinib or temsirolimus in the MCL3001 (RAY) trial
Leukemia (2022)
-
Prognostic value of a novel glycolysis-related gene expression signature for gastrointestinal cancer in the Asian population
Cancer Cell International (2021)
-
Neddylation regulation of mitochondrial structure and functions
Cell & Bioscience (2021)
-
LncRNA ODIR1 inhibits osteogenic differentiation of hUC-MSCs through the FBXO25/H2BK120ub/H3K4me3/OSX axis
Cell Death & Disease (2019)