Transcriptome complexity and riboregulation in the human pathogen Helicobacter pylori
- Research Center for Infectious Diseases, University of Würzburg, Würzburg, Germany
The Gram-negative Epsilonproteobacterium Helicobacter pylori is considered as one of the major human pathogens and many studies have focused on its virulence mechanisms as well as genomic diversity. In contrast, only very little is known about post-transcriptional regulation and small regulatory RNAs (sRNAs) in this spiral-shaped microaerophilic bacterium. Considering the absence of the common RNA chaperone Hfq, which is a key-player in post-transcriptional regulation in enterobacteria, H. pylori was even regarded as an organism without riboregulation. However, analysis of the H. pylori primary transcriptome using RNA-seq revealed a very complex transcriptional output from its small genome. Furthermore, the identification of a wealth of sRNAs as well as massive antisense transcription indicates that H. pylori uses riboregulation for its gene expression control. The ongoing functional characterization of sRNAs along with the identification of associated RNA binding proteins will help to understand their potential roles in Helicobacter virulence and stress response. Moreover, research on riboregulation in H. pylori will provide new insights into its virulence mechanisms and will also help to shed light on post-transcriptional regulation in other Epsilonproteobacteria, including widespread and emerging pathogens such as Campylobacter.
Introduction
For a successful infection of the host or survival in the environment, bacteria have to rapidly adapt their gene expression in response to changing conditions. Besides regulation at the transcriptional level, post-transcriptional regulation is an important layer of gene expression control in both pro- and eukaryotes. Together with several RNA binding proteins which influence RNA structure and stability, the 50- to 400-nt long bacterial small RNAs (sRNAs) act as post-transcriptional regulators under various stress and growth conditions or during virulence (Waters and Storz, 2009; Papenfort and Vogel, 2010; Storz et al., 2011). For example, RyhB and OxyS sRNAs from E. coli are involved in iron metabolism and the oxidative stress response, respectively (Altuvia et al., 1997; Masse and Gottesman, 2002).
Although a small number of sRNAs can directly bind and interfere with protein activity, the majority of functionally characterized sRNAs act as antisense RNAs by base-pairing with target mRNAs (Waters and Storz, 2009). Cis-encoded sRNAs originate from the opposite DNA strand relative to the target and share fully complementary regions with their target genes. In contrast, trans-encoded sRNAs are encoded elsewhere in the genome and interact with their target mRNAs by short and imperfect base-pairing. Most of the trans-encoded sRNAs compete with ribosome binding and, thus, lead to translation repression, which is often coupled to mRNA degradation. Furthermore, an increasing number of sRNAs that activate target-gene expression by various mechanisms have been described (Fröhlich and Vogel, 2009). Similar to transcription factors, sRNAs can modulate the expression of multiple target genes, thereby functioning as key regulators in metabolic pathways or during stress responses (Papenfort and Vogel, 2009; Beisel and Storz, 2010). Besides sRNAs, the RNA chaperone Hfq is a key-player in sRNA-mediated regulation in many bacteria and is required for the stability and activity of sRNAs (Vogel and Luisi, 2011).
A variety of experimental and biocomputational approaches have been developed to identify sRNAs (Altuvia, 2007; Sharma and Vogel, 2009; Backofen and Hess, 2010). Genome-wide computational approaches in particular have predicted hundreds of sRNA candidates in diverse bacteria, which now await experimental validation and functional characterization. Furthermore, massively parallel cDNA-sequencing (RNA-seq) based on next-generation sequencing methods has been revolutionizing transcriptome studies in prokaryotes, including several important pathogens (Wang et al., 2009; Croucher and Thomson, 2010; van Vliet, 2010).
Helicobacter pylori is a Gram-negative, microaerophilic Epsilonproteobacterium which colonizes the stomach of about half of the world’s population, where it can lead to peptic ulcer disease, chronic gastritis, or gastric cancer (Cover and Blaser, 2009). Surprisingly, only a few transcriptional regulators have been discovered in the small 1.67 Mbp H. pylori genome, including only three sigma factors, namely RpoD, FliA, and RpoN (Tomb et al., 1997). In contrast to enterobacteria, almost nothing is known about post-transcriptional regulation in Helicobacter. The systematic identification and subsequent functional characterization of sRNAs will help us to understand the role of riboregulation in H. pylori and provide new insights into its virulence mechanisms.
RNA-seq Reveals a Complex Transcriptome Structure and First sRNAs in H. pylori
None of the enterobacterial sRNAs, except for the housekeeping RNAs, tmRNA, SRP RNA, and M1 RNA, are conserved in H. pylori (Sharma et al., 2010). Until recently, Helicobacter was even regarded as an organism that lacks riboregulation and that carries only basic regulatory circuits, due to its small genome size, a limited number of transcriptional regulators, and the apparent absence of the RNA chaperone Hfq (Mitarai et al., 2007). In addition, based on biocomputational predictions and a small scale cDNA cloning approach only a few natural antisense transcripts and sRNAs were predicted in Helicobacter (Livny et al., 2006; Xiao et al., 2009a,b). However, a novel differential RNA sequencing approach (dRNA-seq) selective for the 5′ end of primary transcripts allowed for the definition of a global map of transcriptional start sites and operons in H. pylori strain 26695, revealing a very complex and compact transcriptome structure (Sharma et al., 2010). Moreover, it led to the discovery of massive antisense transcription as well as an unexpected high number of more than 60 small RNAs including potential regulators of cis- and trans-encoded mRNA targets, indicating that H. pylori uses riboregulation for its gene expression control (Figure 1).
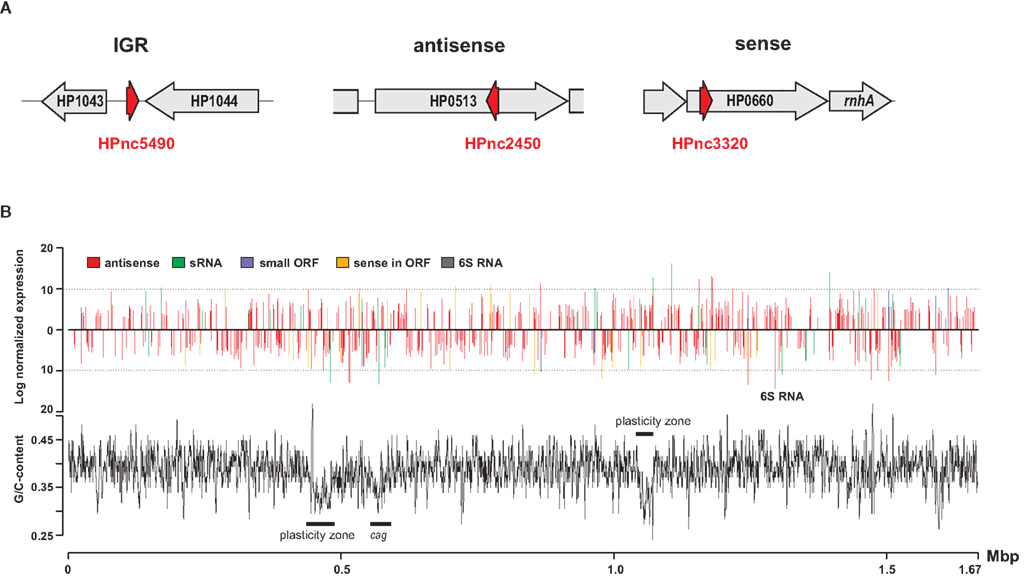
Figure 1. Helicobacter pylori sRNA candidates identified by dRNA-seq. (A) A differential RNA-seq analysis of the primary transcriptome of H. pylori strain 26695 revealed hundreds of novel sRNA candidates (denoted as HPnc). These are located either in intergenic regions (left), antisense to ORFs (middle), or sense to ORFs (right). (B) Relative expression of the different types of sRNA candidates is given as log2 values of read numbers of all sequenced libraries (upper part) and plotted together with local G/C-content (lower part) across the H. pylori strain 26695 genome. One of the most abundant sRNAs is the previously missed 6S RNA homolog from the Epsilon subdivision (gray bar). The Figure was adapted from Sharma et al. (2010).
Potential Functions of sRNAs in H. pylori
Many different in vitro and in vivo approaches have been established to characterize sRNAs and to validate their targets (Vogel and Wagner, 2007; Sharma and Vogel, 2009). Since sRNAs were discovered only recently in Helicobacter our knowledge about their mechanisms, target genes, and their own transcriptional regulators is still limited. The only sRNA that has so far been shown to be essential and required for stress response in H. pylori is tmRNA, which is involved in rescue of stalled ribosomes (Thibonnier et al., 2008).
Cis-encoded sRNAs
Cis-encoded antisense RNAs can overlap the 5′- or 3′-end, the middle, or entire genes and influence expression of their target genes, e.g., by translation inhibition, transcription interference and attenuation, transcript stabilization, or degradation (Thomason and Storz, 2010). Using dRNA-seq, more than 900 cis-encoded antisense RNAs have been identified in H. pylori, including at least one antisense transcriptional start site for almost half of all ORFs (Sharma et al., 2010). Similarly, an increasing number of antisense transcripts have been observed in many other prokaryotes (Thomason and Storz, 2010; Georg and Hess, 2011). Whether all of them are functional or rather represent spurious transcription still needs to be clarified. Obviously, these antisense RNAs can in principle harbor a regulatory function, since expression of an artificial antisense RNA has been successfully used to repress the essential ahpC gene, encoding for alkyl hydroperoxide reductase (Croxen et al., 2007). Since H. pylori lacks homologs of the endonucleolytic RNases E/G (Tomb et al., 1997; Parkhill et al., 2000), antisense-mediated processing by the double-strand specific ribonuclease RNase III could be a major layer of gene regulation in these bacteria, as was recently suggested for Staphylococcus aureus (Lasa et al., 2011).
The characterization of a naturally occurring 292-nt long cis-encoded antisense sRNA from the opposite strand of the urease operon in H. pylori strain 43504 further demonstrated the functionality of antisense RNAs in Helicobacter (Wen et al., 2010). In-vitro and in-vivo approaches demonstrated that 5′ureB–sRNA is induced under neutral pH conditions by the unphosphorylated ArsR response regulator of the acid-responsive ArsRS two-component-system. In its phosphorylated form, ArsR activates transcription of the urease operon in response to low pH (Pflock et al., 2005). The 5′ureB–sRNA shows full complementarity to the 5′ end of ureB mRNA and represses urease activity by mediating cleavage of the ureAB mRNA (Figure 2A). Whether the truncated ureAB transcript is degraded afterwards or whether the reduced amount of UreB leads to lower urease activity is still unclear.
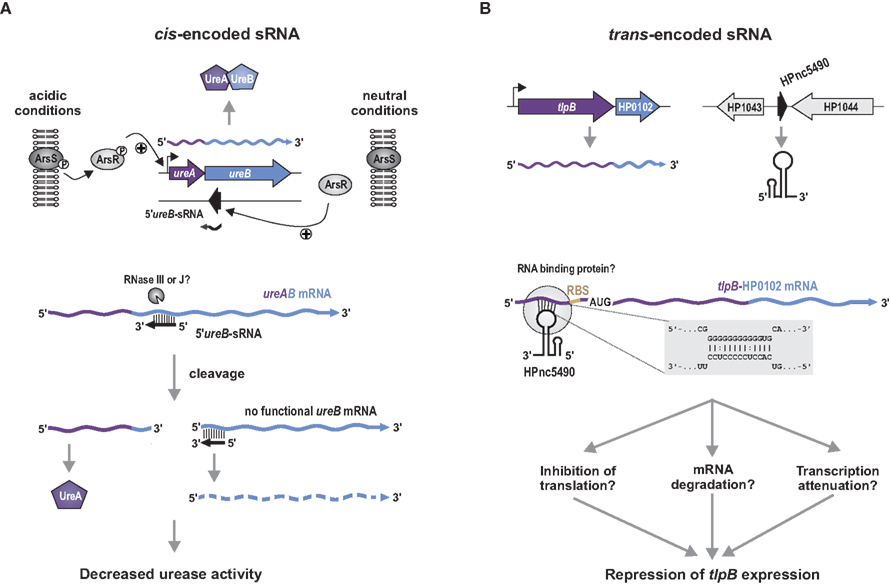
Figure 2. First examples for cis- and trans-encoded antisense RNAs in H. pylori. (A) Regulation of urease by the cis-encoded antisense 5′ureB–sRNA. Expression of the ureAB operon, which encodes for the urease apo-enzyme, is induced under acidic conditions by the phosphorylated ArsR response regulator of the acid-responsive ArsRS two-component-system. In contrast, expression of the cis-encoded antisense transcript, 5′ureB–sRNA, is induced by unphosphorylated ArsR under neutral conditions. An interaction between 5′ureB–sRNA and the 5′ region of ureB leads to cleavage of ureAB mRNA, possible mediated by RNase III or RNase J. Truncation of ureB mRNA prevents its translation, resulting in a reduced amount of UreB and an overall decrease of urease activity. (B) Repression of the chemotaxis receptor TlpB by HPnc5490 sRNA. The 87-nt long HPnc5490 sRNA is encoded in the intergenic region next to the orphan response regulator HP1043. HPnc5490 binds to a G-stretch upstream of the RBS in the 5′ UTR of tlpB–HP0102 mRNA by short and imperfect base-pairing (gray panel), which could be facilitated by a so far unknown RNA binding protein. The exact mechanism of how HPnc5490 mediates repression of tlpB is still unknown but could be mediated either by inhibition of translation, mRNA destabilization by recruitment of RNases, or transcription attenuation.
Besides sRNAs, the dRNA-seq study revealed several new small hydrophobic proteins (<50 aa), some of which are associated with cis-encoded antisense RNAs (Sharma et al., 2010). For example, six structurally related ∼80 nt sRNAs, IsoA1–6 (RNA-inhibitor of small-ORF family A), are expressed antisense to the small ORFs, AapA1–6 (antisense-RNA-associated peptide family A), representing homologous 22–30 aa long peptides. Some of these small ORFs resemble antimicrobial peptides or small toxic peptides from bacteria. The latter are often part of so-called class I toxin–antitoxin loci in which an unstable RNA antitoxin represses expression of a stable peptide toxin. Overexpression of the AapA peptides in H. pylori leads to cell death but not to cell lysis (Darfeuille, personal communication) and in-vitro translation assays indicated that protein synthesis of the peptides is specifically inhibited by the cognate antisense RNAs (Sharma et al., 2010). Thus, the aapA–isoA loci might represent the first examples of class I toxin–antitoxin systems in H. pylori.
Trans-Encoded SRNAs
One of the most abundant transcripts that was identified in the H. pylori dRNA-seq study is the previously missed homolog of 6S RNA (Figure 1B), a ubiquitous riboregulator, which mimics an open promoter complex and thereby sequesters RNA polymerase (Wassarman and Storz, 2000; Barrick et al., 2005; Sharma et al., 2010). Despite only little sequence conservation to E. coli 6S RNA, the 180-nt long RNA from H. pylori can fold into the characteristic long hairpin structure of 6S RNA (Trotochaud and Wassarman, 2005). The dRNA-seq data also revealed 12 to 15–nt long product RNAs, which are transcribed using 6S RNA as a template (Wassarman and Saecker, 2006), providing further evidence that this RNA is a functional 6S homolog in H. pylori. Deletion of 6S RNA results in no obvious phenotype during exponential growth but altered cell survival during stationary phase and under extreme stress conditions in E. coli (Wassarman, 2007). Whether 6S RNA has a role during stress response or stationary phase growth in H. pylori or, like in Legionella (Faucher et al., 2010), impacts on its virulence still needs to be investigated.
Several of the newly identified sRNAs in Helicobacter are potential candidates for trans-encoded antisense RNAs. For example, the abundant 87-nt long HPnc5490 sRNA, was predicted to interact by a C/U rich stretch with a G-repeat in the 5′ UTR of tlpB mRNA encoding for one of the four chemotaxis receptors in H. pylori (Figure 2B; Sharma et al., 2010). Comparison of tlpB expression in the wild-type and a HPnc5490 deletion strain confirmed down-regulation of the tlpB mRNA as well as TlpB protein levels by HPnc5490. It has been suggested that TlpB senses protons and diverse studies have demonstrated its potential role in pH-taxis, quorum sensing as well as colonization, and inflammation of the gastric mucosa (McGee et al., 2005; Croxen et al., 2006; Williams et al., 2007; Rader et al., 2011). Therefore, HPnc5490, and probably additional H. pylori sRNAs, could play important roles during stress responses or infection, as described for other bacterial pathogens (Papenfort and Vogel, 2010).
Protein Factors Involved in Post-Transcriptional Regulation
Besides sRNAs, RNA binding proteins and RNases are involved in post-transcriptional regulation in bacteria (Pichon and Felden, 2007). In H. pylori, analysis of the urease operon transcript revealed different mRNA stabilities dependent on the pH value and processing into multiple species, indicating an extensive post-transcriptional regulation of the urease mRNA (Akada et al., 2000). Moreover, it has been shown that a gene of previously unknown function, HP0958, is essential for flagellum biogenesis and encodes for a post-transcriptional regulator of flagellin mRNA by modulating the amount of flaA mRNA available for translation (Douillard et al., 2008). Together with the recent identification of sRNAs it is now clear that H. pylori uses post-transcriptional regulation to control its gene expression and future identification of the protein factors involved will help to understand the underlying mechanisms in these bacteria.
RNA Binding Proteins
The Sm-like RNA chaperone Hfq is required for the stabilization of sRNAs and facilitates the base-pairing of trans-encoded sRNA with their target mRNAs (Vogel and Luisi, 2011). Deletion of hfq leads to pleiotropic phenotypes including reduced fitness and virulence in several bacterial pathogens (Chao and Vogel, 2010). However, as is the case for 50% of all bacterial genomes, hfq appears to be absent in Epsilonproteobacteria. Whether a different RNA binding protein replaces the function of Hfq or whether sRNAs act without a chaperone in these bacteria is still an open question.
Besides Hfq, several other RNA binding proteins, such as the carbon storage regulator CsrA/RsmA family or the Crc protein involved in catabolite repression participate in post-transcriptional regulation in bacteria (Sonnleitner et al., 2009; Timmermans and Van Melderen, 2010). The CsrA protein acts as a translation regulator and its own activity is regulated by two sRNAs, CsrB/C, which mimic the RNA substrate of CsrA (Babitzke and Romeo, 2007). The CsrA homolog in Helicobacter is required for full motility, survival under oxidative stress, and infection of mice (Barnard et al., 2004). However, neither the target genes of CsrA nor the associated CsrA-regulating sRNAs, CsrB/C, have been identified in H. pylori, yet. The recent findings that additional proteins with so far unknown functions or functions unrelated to RNA metabolism, are involved in post-transcriptional control of mRNAs are just one indication that we are far away from knowing all RNA binding proteins and their roles in bacteria (Mitobe et al., 2011; Pandey et al., 2011).
Ribonucleases
Although the exact mechanism of repression is still unclear, both functionally characterized antisense RNAs in Helicobacter (Figure 2) cause reduced protein levels as well as reduced target-mRNA levels, most likely through transcript destabilization or active recruitment of RNases. In enterobacteria, sRNA-mediated target-mRNA decay mainly depends on the RNA degradosome, a protein complex composed of the endoribonuclease RNase E, polynucleotide phosphorylase PNPase, and the RNA helicase RhlB (Morita et al., 2005; Caron et al., 2010). In H. pylori, several RNases are annotated (RNase H, RNase H-II, RNase J, RNase N, RNase P, RNase R, and RNase III) but like all Epsilonproteobacteria it appears to lack a homolog of RNase E. Recent studies on Bacillus subtilis and S. aureus identified RNase J1 and RNAse J2 as the functional orthologs in the RNA degradosome of Gram-positive bacteria (Mathy et al., 2010; Roux et al., 2011). Furthermore, in S. aureus the double-strand specific RNase III degrades, mainly in concert with the sRNA RNAIII, several mRNAs encoding virulence factors (Huntzinger et al., 2005; Boisset et al., 2007; Chevalier et al., 2008). Therefore, RNase J and RNase III could be potential RNases participating in sRNA-mediated transcript destabilization in H. pylori. Moreover, the 3′–5′ exoribonuclease RNase R has been shown to post-transcriptionally down-regulate six virulence related genes (Tsao et al., 2009), but its potential role in sRNA-mediated regulation still needs to be investigated.
Transcriptional Regulation of Small RNAs in H. pylori
In enterobacteria, several of the functionally characterized sRNAs have been shown be key regulators of larger regulatory networks (Papenfort and Vogel, 2009; Beisel and Storz, 2010). Moreover, some sRNAs are controlled by key transcriptional regulators of certain metabolic pathways and several transcriptional regulators have been shown to be themselves regulated at the post-transcriptional level (Storz et al., 2011). Thus, besides the identification of their target genes, the investigation of transcriptional regulators of sRNAs and antisense RNAs will help provide insights in the conditions in which their expression is required and what are their physiological roles. As the H. pylori genome contains only a few transcriptional regulators, it is likely that some of the sRNAs are regulated by one or more of these transcription factors and vice versa.
Perspective
The discovery of an unexpected wealth of sRNAs in Helicobacter opens an additional area of gene expression control in this pathogenic Epsilonproteobacterium. The characterization of selected cis- and trans-encoded sRNAs will uncover their potential regulatory functions in virulence and stress response of H. pylori. Moreover, these studies could reveal novel mechanisms of post-transcriptional regulation independent of the RNA chaperone Hfq. The identification of novel ribonucleoprotein complexes will expand our knowledge about cellular regulators and could provide new targets for antimicrobial therapies against emerging pathogens.
Besides mechanistic aspects of riboregulation, research on sRNAs in H. pylori could also help to understand phenotypes observed in previous genetic screens. Global transposon screens failed to hit around 340 genes indicating their potential essentiality and revealed about 220 candidate mutants with a colonization defect in mice (Salama et al., 2004; Baldwin et al., 2007). Since at least 60 sRNAs have been validated in H. pylori and considering the high fraction of cis-encoded antisense RNAs, several of the virulence associated genes could be subjected to post-transcriptional regulation. For example, the above mentioned first examples of functional sRNAs in H. pylori prove post-transcriptional regulation of the virulence factors, urease, and TlpB in H. pylori (Sharma et al., 2010; Wen et al., 2010). In addition, a transposon screen for virulence and colonization factors in H. pylori revealed several mutants with a transposon hit at the end of genes or in intergenic regions, indicating that there might be additional sRNAs with a role in virulence (Baldwin et al., 2007).
Next-generation sequencing methods have turned out to be an effective tool to uncover the transcriptome structure and identify novel transcripts in prokaryotes. RNA-seq based expression profiling of bacteria grown under different conditions or in comparison to mutant strains will help to identify sRNAs, which are induced under certain stress or virulence conditions. Moreover, the full understanding of infection processes requires the parallel investigation of gene expression in both, pathogen and host. Microarray-based techniques are limited by the requirement of relatively high amounts of starting material as well as cross-hybridization problems of host and pathogen RNA. RNA-seq methods could overcome those limitations by parallel sequencing of host and pathogen RNA without any prior separation steps followed by mapping of the cDNA reads to the host and pathogen genomes. Furthermore, the investigation of single cell transcriptomes (Tang et al., 2009; Kang et al., 2011) as well as the study of expression profiles of several strains by comparative RNA-seq approaches could lead to new insights into gene expression in bacterial sub-populations or differences between pathogenic and non-pathogenic species.
The study of riboregulation in H. pylori will also help to understand post-transcriptional regulation in other Epsilonproteobacteria, such as the emerging pathogen Campylobacter jejuni, which is considered as the leading cause of bacterial gastroenteritis (Young et al., 2007). Recently, first sRNA candidates have also been identified in C. jejuni NCTC11168 based on conventional RNA-seq combined with predictions of conserved RNA structures (Chaudhuri et al., 2011). A dRNA-seq analysis of the same strain revealed more than 20 candidate sRNAs as well as a complex and compact transcriptional output (Porcelli and van Vliet, personal communication). A comparative dRNA-seq analysis of multiple C. jejuni strains indicates that many of the sRNAs are conserved among C. jejuni strains and show similar expression profiles (Dugar and Sharma, unpublished). However, some conserved sRNAs show distinct expression patterns in certain strains indicating that they could have varying function. Overall, Helicobacter and Campylobacter seem to have evolved their own specific sRNA repertoires, but with respect to their smaller genome sizes compared to E. coli express a comparable number of sRNAs. Research on sRNAs in Helicobacter and Campylobacter as well as parallel transcriptome analysis of host and pathogen during the time-course of infection will help to shed light on post-transcriptional regulation and virulence mechanisms not only in Epsilonproteobacteria, but also other bacterial pathogens, including species that lack Hfq.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This publication was funded by the German Research Foundation (DFG) and the University of Würzburg in the funding program Open Access Publishing. We thank Fabien Darfeuille, Kathrin Fröhlich, Kai Papenfort, Colin Corcoran, and Alexander Böhm for critical comments on this manuscript.
References
Akada, J. K., Shirai, M., Takeuchi, H., Tsuda, M., and Nakazawa, T. (2000). Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36, 1071–1084.
Altuvia, S. (2007). Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10, 257–261.
Altuvia, S., Weinstein-Fischer, D., Zhang, A., Postow, L., and Storz, G. (1997). A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90, 43–53.
Babitzke, P., and Romeo, T. (2007). CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10, 156–163.
Backofen, R., and Hess, W. R. (2010). Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 7, 33–42.
Baldwin, D. N., Shepherd, B., Kraemer, P., Hall, M. K., Sycuro, L. K., Pinto-Santini, D. M., and Salama, N. R. (2007). Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect. Immun. 75, 1005–1016.
Barnard, F. M., Loughlin, M. F., Fainberg, H. P., Messenger, M. P., Ussery, D. W., Williams, P., and Jenks, P. J. (2004). Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 51, 15–32.
Barrick, J. E., Sudarsan, N., Weinberg, Z., Ruzzo, W. L., and Breaker, R. R. (2005). 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11, 774–784.
Beisel, C. L., and Storz, G. (2010). Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 34, 866–882.
Boisset, S., Geissmann, T., Huntzinger, E., Fechter, P., Bendridi, N., Possedko, M., Chevalier, C., Helfer, A. C., Benito, Y., Jacquier, A., Gaspin, C., Vandenesch, F., and Romby, P. (2007). Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21, 1353–1366.
Caron, M. P., Lafontaine, D. A., and Masse, E. (2010). Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 7, 140–144.
Chao, Y., and Vogel, J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33.
Chaudhuri, R. R., Yu, L., Kanji, A., Perkins, T. T., Gardner, P. P., Choudhary, J., Maskell, D. J., and Grant, A. J. (2011). Quantitative RNA-seq analysis of the transcriptome of Campylobacter jejuni. Microbiology 157, 2922–2932.
Chevalier, C., Huntzinger, E., Fechter, P., Boisset, S., Vandenesch, F., Romby, P., and Geissmann, T. (2008). Staphylococcus aureus endoribonuclease III purification and properties. Meth. Enzymol. 447, 309–327.
Cover, T. L., and Blaser, M. J. (2009). Helicobacter pylori in health and disease. Gastroenterology 136, 1863–1873.
Croucher, N. J., and Thomson, N. R. (2010). Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 13, 619–624.
Croxen, M. A., Ernst, P. B., and Hoffman, P. S. (2007). Antisense RNA modulation of alkyl hydroperoxide reductase levels in Helicobacter pylori correlates with organic peroxide toxicity but not infectivity. J. Bacteriol. 189, 3359–3368.
Croxen, M. A., Sisson, G., Melano, R., and Hoffman, P. S. (2006). The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188, 2656–2665.
Douillard, F. P., Ryan, K. A., Caly, D. L., Hinds, J., Witney, A. A., Husain, S. E., and O’Toole, P. W. (2008). Posttranscriptional regulation of flagellin synthesis in Helicobacter pylori by the RpoN chaperone HP0958. J. Bacteriol. 190, 7975–7984.
Faucher, S. P., Friedlander, G., Livny, J., Margalit, H., and Shuman, H. A. (2010). Legionella pneumophila 6S RNA optimizes intracellular multiplication. Proc. Natl. Acad. Sci. U.S.A. 107, 7533–7538.
Fröhlich, K. S., and Vogel, J. (2009). Activation of gene expression by small RNA. Curr. Opin. Microbiol. 12, 674–682.
Georg, J., and Hess, W. R. (2011). cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75, 286–300.
Huntzinger, E., Boisset, S., Saveanu, C., Benito, Y., Geissmann, T., Namane, A., Lina, G., Etienne, J., Ehresmann, B., Ehresmann, C., Jacquier, A., Vandenesch, F., and Romby, P. (2005). Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24, 824–835.
Kang, Y., Norris, M. H., Zarzycki-Siek, J., Nierman, W. C., Donachie, S. P., and Hoang, T. T. (2011). Transcript amplification from single bacterium for transcriptome analysis. Genome Res. 21, 925–935.
Lasa, I., Toledo-Arana, A., Dobin, A., Villanueva, M., de Los Mozos, I. R., Vergara-Irigaray, M., Segura, V., Fagegaltier, D., Penades, J. R., Valle, J., Solano, C., and Gingeras, T. R. (2011). Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 20172–20177.
Livny, J., Brencic, A., Lory, S., and Waldor, M. K. (2006). Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 34, 3484–3493.
Masse, E., and Gottesman, S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625.
Mathy, N., Hebert, A., Mervelet, P., Benard, L., Dorleans, A., Li de la Sierra-Gallay, I., Noirot, P., Putzer, H., and Condon, C. (2010). Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol. Microbiol. 75, 489–498.
McGee, D. J., Langford, M. L., Watson, E. L., Carter, J. E., Chen, Y. T., and Ottemann, K. M. (2005). Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73, 1820–1827.
Mitarai, N., Andersson, A. M., Krishna, S., Semsey, S., and Sneppen, K. (2007). Efficient degradation and expression prioritization with small RNAs. Phys. Biol. 4, 164–171.
Mitobe, J., Yanagihara, I., Ohnishi, K., Yamamoto, S., Ohnishi, M., Ishihama, A., and Watanabe, H. (2011). RodZ regulates the post-transcriptional processing of the Shigella sonnei type III secretion system. EMBO Rep. 12, 911–916.
Morita, T., Maki, K., and Aiba, H. (2005). RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19, 2176–2186.
Pandey, S. P., Minesinger, B. K., Kumar, J., and Walker, G. C. (2011). A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 39, 4691–4708.
Papenfort, K., and Vogel, J. (2009). Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 160, 278–287.
Papenfort, K., and Vogel, J. (2010). Regulatory RNA in bacterial pathogens. Cell Host Microbe 8, 116–127.
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., Chillingworth, T., Davies, R. M., Feltwell, T., Holroyd, S., Jagels, K., Karlyshev, A. V., Moule, S., Pallen, M. J., Penn, C. W., Quail, M. A., Rajandream, M. A., Rutherford, K. M., van Vliet, A. H., Whitehead, S., and Barrell, B. G. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668.
Pflock, M., Kennard, S., Delany, I., Scarlato, V., and Beier, D. (2005). Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73, 6437–6445.
Pichon, C., and Felden, B. (2007). Proteins that interact with bacterial small RNA regulators. FEMS Microbiol. Rev. 31, 614–625.
Rader, B. A., Wreden, C., Hicks, K. G., Sweeney, E. G., Ottemann, K. M., and Guillemin, K. (2011). Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157, 2445–2455.
Roux, C. M., DeMuth, J. P., and Dunman, P. M. (2011). Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J. Bacteriol. 193, 5520–5526.
Salama, N. R., Shepherd, B., and Falkow, S. (2004). Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186, 7926–7935.
Sharma, C. M., Hoffmann, S., Darfeuille, F., Reignier, J., Findeiss, S., Sittka, A., Chabas, S., Reiche, K., Hackermuller, J., Reinhardt, R., Stadler, P. F., and Vogel, J. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255.
Sharma, C. M., and Vogel, J. (2009). Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 12, 536–546.
Sonnleitner, E., Abdou, L., and Haas, D. (2009). Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 106, 21866–21871.
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891.
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., Wang, X., Bodeau, J., Tuch, B. B., Siddiqui, A., Lao, K., and Surani, M. A. (2009). mRNA-seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382.
Thibonnier, M., Thiberge, J. M., and De Reuse, H. (2008). Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS ONE 3:e3810. doi:10.1371/journal.pone.0003810
Thomason, M. K., and Storz, G. (2010). Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 44, 167–188.
Timmermans, J., and Van Melderen, L. (2010). Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci. 67, 2897–2908.
Tomb, J. F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., Nelson, K., Quackenbush, J., Zhou, L., Kirkness, E. F., Peterson, S., Loftus, B., Richardson, D., Dodson, R., Khalak, H. G., Glodek, A., McKenney, K., Fitzegerald, L. M., Lee, N., Adams, M. D., Hickey, E. K., Berg, D. E., Gocayne, J. D., Utterback, T. R., Peterson, J. D., Kelley, J. M., Cotton, M. D., Weidman, J. M., Fujii, C., Bowman, C., Watthey, L., Wallin, E., Hayes, W. S., Borodovsky, M., Karp, P. D., Smith, H. O., Fraser, C. M., and Venter, J. C. (1997). The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547.
Trotochaud, A. E., and Wassarman, K. M. (2005). A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 12, 313–319.
Tsao, M. Y., Lin, T. L., Hsieh, P. F., and Wang, J. T. (2009). The 3′-to-5′ exoribonuclease (encoded by HP1248) of Helicobacter pylori regulates motility and apoptosis-inducing genes. J. Bacteriol. 191, 2691–2702.
van Vliet, A. H. (2010). Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol. Lett. 302, 1–7.
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589.
Vogel, J., and Wagner, E. G. (2007). Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 10, 262–270.
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63.
Wassarman, K. M., and Saecker, R. M. (2006). Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 314, 1601–1603.
Wassarman, K. M., and Storz, G. (2000). 6S RNA regulates E. coli RNA polymerase activity. Cell 101, 613–623.
Wen, Y., Feng, J., Scott, D. R., Marcus, E. A., and Sachs, G. (2010). A cis-encoded antisense small RNA regulated by the HP0165-HP0166 two-component system controls expression of ureB in Helicobacter pylori. J. Bacteriol. 193, 40–51.
Williams, S. M., Chen, Y. T., Andermann, T. M., Carter, J. E., McGee, D. J., and Ottemann, K. M. (2007). Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75, 3747–3757.
Xiao, B., Li, W., Guo, G., Li, B., Liu, Z., Jia, K., Guo, Y., Mao, X., and Zou, Q. (2009a). Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Curr. Microbiol. 58, 258–263.
Xiao, B., Li, W., Guo, G., Li, B. S., Liu, Z., Tang, B., Mao, X. H., and Zou, Q. M. (2009b). Screening and identification of natural antisense transcripts in Helicobacter pylori by a novel approach based on RNase I protection assay. Mol. Biol. Rep. 36, 1853–1858.
Keywords: RNA-seq, sRNA, Helicobacter pylori, post-transcriptional regulation, transcriptome analysis
Citation: Pernitzsch SR and Sharma CM (2012) Transcriptome complexity and riboregulation in the human pathogen Helicobacter pylori. Front. Cell. Inf. Microbio. 2:14. doi: 10.3389/fcimb.2012.00014
Received: 15 December 2011; Accepted: 02 February 2012;
Published online: 21 February 2012.
Edited by:
D. Scott Merrell, Uniformed Services University, USAReviewed by:
D. Scott Merrell, Uniformed Services University, USAGisela Storz, National Institutes of Health, USA
David Scott, David Geffen School of Medicine at UCLA, USA
Copyright: © 2012 Pernitzsch and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Cynthia M. Sharma, Research Center for Infectious Diseases (ZINF), University of Würzburg, Josef-Schneider-Str. 2/Bau D15, 97080 Würzburg, Germany. e-mail: cynthia.sharma@uni-wuerzburg.de