Molecular Networking-Based Analysis of Cytotoxic Saponins from Sea Cucumber Holothuria atra
Abstract
:1. Introduction
2. Results
2.1. Saponin Profile and MS/MS-Based Molecular Networking
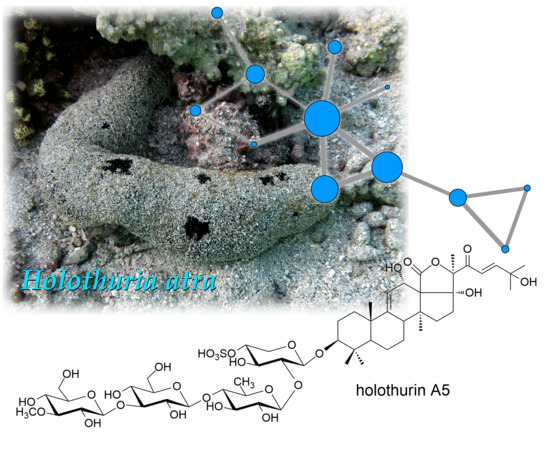
2.2. Structural Determination of Holothurin A5 (1)
2.3. Solvent Addition to Holothurin A5 (1) and Structure of Compounds 5 and 6
2.4. Biological Activity of Compounds 1–4
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Sample Collection, Extraction, and Isolation
4.3. LC-HRMS and LC-HRMS/MS
4.4. Molecular Networking
4.5. In Vitro Cytotoxic Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shakouri, A.; Nabavi, M.B.; Kochanian, P.; Savari, A.; Safahieh, A.; Aminrad, T. New observation of two species of sea cucumbers from Chabahar bay (Southeast coasts of Iran). Asian J. Anim. Sci. 2009, 3, 130–134. [Google Scholar] [CrossRef]
- Khotimchenko, Y. Pharmacological potential of sea cucumbers. Int. J. Mol. Sci. 2018, 19, 1342. [Google Scholar] [CrossRef] [PubMed]
- Janakiram, N.B.; Mohammed, A.; Rao, C.V. Sea cucumbers metabolites as potent anti-cancer agents. Mar. Drugs 2015, 13, 2909–2923. [Google Scholar] [CrossRef] [PubMed]
- Haug, T.; Kjuul, A.K.; Styrvold, O.B.; Sandsdalen, E.; Olsen, Ø.M.; Stensvåg, K. Antibacterial activity in Strongylocentrotus droebachiensis (Echinoidea), Cucumaria frondosa (Holothuroidea), and Asterias rubens (Asteroidea). J. Invertebr. Pathol. 2002, 81, 94–102. [Google Scholar] [CrossRef]
- Mashjoor, S.; Yousefzadi, M. Holothurians antifungal and antibacterial activity to human pathogens in the Persian Gulf. J. Mycol. Med. 2017, 27, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.H.; Yi, Y.H.; Tang, H.F.; Liu, B.S.; Wang, Z.L.; Sun, G.Q.; Zhang, W.; Li, L.; Sun, P. Antifungal triterpene glycosides from the sea cucumber Bohadschia marmorata. Planta Med. 2009, 75, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Farshadpour, F.; Gharibi, S.; Taherzadeh, M.; Amirinejad, R.; Taherkhani, R.; Habibian, A.; Zandi, K. Antiviral activity of Holothuria sp. a sea cucumber against herpes simplex virus type 1 (HSV-1). Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 333–337. [Google Scholar] [PubMed]
- Mena-Bueno, S.; Atanasova, M.; Fernández-Trasancos, Á.; Paradela-Dobarro, B.; Bravo, S.B.; Álvarez, E.; Fernández, Á.L.; Carrera, I.; González-Juanatey, J.R.; Eiras, S. Sea cucumbers with an anti-inflammatory effect on endothelial cells and subcutaneous but not on epicardial adipose tissue. Food Funct. 2016, 7, 953–963. [Google Scholar] [CrossRef]
- Wu, M.; Wen, D.; Gao, N.; Xiao, C.; Yang, L.; Xu, L.; Lian, W.; Peng, W.; Jiang, J.; Zhao, J. Anticoagulant and antithrombotic evaluation of native fucosylated chondroitin sulfates and their derivatives as selective inhibitors of intrinsic factor Xase. Eur. J. Med. Chem. 2015, 92, 257–269. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Z.; Zhang, M.; Meng, X.; Xia, X.; Yuan, W.; Xue, F.; Liu, C. Antioxidant and antihyperlipidemic activities of polysaccharides from sea cucumber Apostichopus japonicus. Carbohydr. Polym. 2012, 90, 1664–1670. [Google Scholar] [CrossRef]
- Siahaan, E.; Pangestuti, R.; Munandar, H.; Kim, S.-K. Cosmeceuticals Properties of Sea Cucumbers: Prospects and Trends. Cosmetics 2017, 4, 26. [Google Scholar] [CrossRef]
- Suleria, H.A.R.; Osborne, S.; Masci, P.; Gobe, G. Marine-based nutraceuticals: An innovative trend in the food and supplement industries. Mar. Drugs 2015, 13, 6336–6351. [Google Scholar] [CrossRef] [PubMed]
- Bordbar, S.; Anwar, F.; Saari, N. High-value components and bioactives from sea cucumbers for functional foods—A review. Mar. Drugs 2011, 9, 1761–1805. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Hori, M.; Kan, K.; Yasuzawa, T.; Matsui, M.; Suzuki, S.; Kitagawa, I. Marine natural products. XXVII. Distribution of lanostane-type triterpene oligoglycosides in ten kinds of Okinawan Sea cucumbers. Chem. Pharm. Bull. 1991, 39, 2282–2287. [Google Scholar] [CrossRef]
- Kalinin, V.I. System-theoretical (Holistic) approach to the modelling of structural-functional relationships of biomolecules and their evolution: An example of triterpene glycosides from sea cucumbers (Echinodermata, Holothurioidea). J. Theor. Biol. 2000, 206, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Van Dyck, S.; Caulier, G.; Todesco, M.; Gerbaux, P.; Fournier, I.; Wisztorski, M.; Flammang, P. The triterpene glycosides of Holothuria forskali: Usefulness and efficiency as a chemical defense mechanism against predatory fish. J. Exp. Biol. 2011, 214, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Caulier, G.; Flammang, P.; Gerbaux, P.; Eeckhaut, I. When a repellent becomes an attractant: Harmful saponins are kairomones attracting the symbiotic Harlequin crab. Sci. Rep. 2013, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Oleinikova, G.K.; Kuznetsova, T.A. Glycosides ofthe holothurian Holothuria atra. Chem. Nat. Compd. 1986, 22, 617. [Google Scholar] [CrossRef]
- Van Dyck, S.; Gerbaux, P.; Flammang, P. Qualitative and quantitative saponin contents in five sea cucumbers from the Indian ocean. Mar. Drugs 2010, 8, 173–189. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Kupchan, S.M.; Britton, R.W.; Ziegler, M.F.; Sioel, C.W. Bruceantin, a New Potent Antileukemic Simaroubolide from Brucea antidysenterica. J. Org. Chem. 1973, 38, 178–179. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitagawa, I.; Nishino, T.; Kobayashi, M.; Matsuno, T.; Akutsu, H.; Kyogoku, Y. Marine natural products. VII. Bioactive triterpene-oligoglycosides from the sea cucumber Holothuria leucospilota Brandt (1). Structure of holothurin B. Chem. Pharm. Bull. 1981, 29, 1942–1950. [Google Scholar] [CrossRef]
- Kitagawa, I.; Inamoto, T.; Fuchida, M.; Okada, S.; Kobayashi, M.; Nishino, T.; Kyogoku, Y. Structures of echinoside A and B, two antifungal oligoglycosides from the sea cucumber Actinopyga echinites (Jaeger). Chem. Pharm. Bull. 1980, 28, 1651–1653. [Google Scholar] [CrossRef]
- Kitagawa, I.; Nishino, T.; Kobayashi, M.; Kyogoku, Y. Marine natural products. VIII. Bioactive triterpene-oligoglycosides from the sea cucumber Holothuria leucospilota Brandt (2). Structure of holothurin A. Chem. Pharm. Bull. 1981, 29, 1951–1956. [Google Scholar] [CrossRef]
- Dang, N.H.; Thanh, N.; Van Kiem, P.; Van Huong, L.M.; Minh, C.; Van Kim, Y.H. Two new triterpene glycosides from the Vietnamese sea cucumber Holothuria scabra. Arch. Pharm. Res. 2007, 30, 1387–1391. [Google Scholar] [CrossRef]
- Xu, X.; Bai, H.; Zhou, L.; Deng, Z.; Zhong, H.; Wu, Z.; Yao, Q. Three new cucurbitane triterpenoids from Hemsleya penxianensis and their cytotoxic activities. Bioorganic Med. Chem. Lett. 2014, 24, 2159–2162. [Google Scholar] [CrossRef]
- Zhu, N.; Sun, Z.; Hu, M.; Li, Y.; Zhang, D.; Wu, H.; Tian, Y.; Li, P.; Yang, J.; Ma, G.; et al. Cucurbitane-type triterpenes from the tubers of Hemsleya penxianensis and their bioactive activity. Phytochemistry 2018, 147, 49–56. [Google Scholar] [CrossRef]
- Kicha, A.A.; Kalinovsky, A.I.; Ivanchina, N.V.; Malyarenko, T.V.; Dmitrenok, P.S.; Kuzmich, A.S.; Sokolova, E.V.; Stonik, V.A. Furostane Series Asterosaponins and Other Unusual Steroid Oligoglycosides from the Tropical Starfish Pentaceraster regulus. J. Nat. Prod. 2017, 80, 2761–2770. [Google Scholar] [CrossRef]
- Moosmann, P.; Ueoka, R.; Grauso, L.; Mangoni, A.; Morinaka, B.I.; Gugger, M.; Piel, J. Cyanobacterial ent-Sterol-Like Natural Products from a Deviated Ubiquinone Pathway. Angew. Chem. Int. Ed. 2017, 56, 4987–4990. [Google Scholar] [CrossRef] [PubMed]





| Measured m/z | Retention Time (min) | Molecular Formula | Putative Identification |
|---|---|---|---|
| 859.38 | 23.8 | C41H63O17S− | holothurin B |
| 1165.51 | 23.1 | C54H85O25S− | unknown |
| 1167.53 | 23.6 | C54H87O25S− | unknown |
| 1181.51 | 23.2 | C54H85O26S− | 24-dehydroechinoside A (4) |
| 1183.52 | 23.7 | C54H87O26S− | echinoside A (3) |
| 1195.48 | 21.5 | C54H83O27S− | calcigeroside B |
| 1197.50 | 21.8 | C54H85O27S− | holothurin A (2) |
| 1211.48 | 19.6 | C54H83O28S− | unknown before this study (1) |
| 1213.50 | 18.3 | C54H85O28S− | holothurin D |
| 1225.50 | 21.2 | C55H85O28S− | unknown |
| 1229.49 | 17.6, 18.1 | C54H85O29S− | unknown before this study (5) |
| 1243.51 | 19.5, 19.7 | C55H87O29S− | unknown before this study (6) |
| Position | δC, Type | δH (Mult, J in Hz) | Sugar | Position | δC, Type | δH (Mult, J in Hz) |
|---|---|---|---|---|---|---|
| 1 | 37.2 (CH2) | 1.51 (m), 1.85 (m) | Xyl | 1′ | 105.5 (CH) | 4.43 (d, 7.5) |
| 2 | 27.4 (CH2) | 1.79 (m), 1.97 (m) | 2′ | 82.2 (CH) | 3.56 (dd, 8.9, 7.5) | |
| 3 | 90.0 (CH) | 3.13 (br. d, 12.0) | 3′ | 75.5 (CH) | 3.73 (t, 8.9) | |
| 4 | 40.9 (C) | 4′ | 77.1 (CH) | 4.22 (m) | ||
| 5 | 53.5 (CH) | 0.98 (d, 12.4) | 5′ | 64.0 (CH2) | 3.37 (t, 10.1), 4.20 (m) | |
| 6 | 21.8 (CH2) | 1.57 (m), 1.76 (m) | ||||
| 7 | 28.8 (CH2) | 1.46 (m), 1.77 (m) | Qui | 1′′ | 104.9 (CH) | 4.61 (d, 7.6) |
| 8 | 41.6 (CH) | 3.02 (dd, 4.0, 13.2) | 2′′ | 76.2 (CH) | 3.29 (dd, 9.0, 7.6) | |
| 9 | 156.2 (C) | 3′′ | 75.6 (CH) | 3.47 (t, 9.0) | ||
| 10 | 40.7 (C) | 4′′ | 86.5 (CH) | 3.17 (t, 9.0) | ||
| 11 | 115.2 (CH) | 5.39 (br. d, 5.7) | 5′′ | 72.2 (CH) | 3.46 (m) | |
| 12 | 71.6 (CH) | 4.55 (br. d, 5.7) | 6′′ | 17.8 (CH3) | 1.36 (d, 6.1) | |
| 13 | 60.0 (C) | |||||
| 14 | 47.3 (C) | Glc | 1′′′ | 104.4 (CH) | 4.42 (d, 7.9) | |
| 15 | 37.2 (CH2) | 1.21 (m), 1.79 (m) | 2′′′ | 74.2 (CH) | 3.41 (m) | |
| 16 | 39.2 (CH2) | 1.90 (m), 2.16 (m) | 3′′′ | 87.2 (CH) | 3.57 (t, 8.9) | |
| 17 | 88.5 (C) | 4′′′ | 69.5 (CH) | 3.41 (m) | ||
| 18 | 175.0 (C) | 5′′′ | 77.4 (CH) | 3.39 (m) | ||
| 19 | 22.6 (CH3) | 1.16 (s) | 6′′′ | 62.3 (CH2) | 3.67 (dd, 11.9, 5.7), 3.89 (dd, 11.9, 2.1) | |
| 20 | 93.0 (C) | |||||
| 21 | 21.1 (CH3) | 1.67 (s) | OMeGlc | 1′′′′ | 104.9 (CH) | 4.58 (d, 7.3) |
| 22 | 198.3 (C) | 2′′′′ | 75.1 (CH) | 3.32 (m) | ||
| 23 | 121.3 (CH) | 6.85 (d, 15.9) | 3′′′′ | 87.3 (CH) | 3.11 (t, 8.7) | |
| 24 | 157.6 (CH) | 7.14 (d, 15.9) | 4′′′′ | 70.8 (CH) | 3.33 (m) | |
| 25 | 71.9 (C) | 5′′′′ | 77.8 (CH) | 3.33 (m) | ||
| 26 | 28.8 (CH3) | 1.34 (s) | 6′′′′ | 62.5 (CH2) | 3.64 (dd, 11.7, 5.7), 3.86 (dd, 11.7, 1.7) | |
| 27 | 28.8 (CH3) | 1.34 (s) | OMe | 60.8 (CH3) | 3.63 (s) | |
| 30 | 16.9 (CH3) | 0.92 (s) | ||||
| 31 | 28.3 (CH3) | 1.09 (s) | ||||
| 32 | 19.8 (CH3) | 1.29 (s) |
| Position | Proton Count | Compound 5 | Compound 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 5a | 5b | 6a | 6b | ||||||
| δC | δH | δC | δH | δC | δH | δC | δH | ||
| 17 | C | 88.1 | - | 88.4 | - | 88.1 | 88.4 | ||
| 18 | C | 175.0 | - | 175.0 | - | 175.0 | 175.0 | ||
| 19 | CH3 | 22.6 | 1.16 | 22.6 | 1.16 | 22.6 | 1.16 | 22.6 | 1.16 |
| 20 | C | 93.0 | 93.0 | 92.6 | 93.2 | ||||
| 21 | CH3 | 21.1 | 1.674 | 21.1 | 1.667 | 21.1 | 1.672 | 21.1 | 1.669 |
| 22 | C | 209.8 | 209.7 | 209.1 | 209.7 | ||||
| 23 | CH2 | 41.8 | 2.83, 2.86 | 41.8 | 2.83, 2.86 | 41.8 | 2.88, 2.92 | 41.8 | 2.88, 2.92 |
| 24 | CH | 73.9 | 3.95 | 74.2 | 3.95 | 84.4 | 3.64 | 84.0 | 3.64 |
| 25 | C | 73.1 | 73.2 | 73.5 | 73.6 | ||||
| 26 | CH3 | 24.1 | 1.152 | 24.2 | 1.156 | 24.6 | 1.136 | 24.8 | 1.144 |
| 27 | CH3 | 26.8 | 1.218 | 26.7 | 1.208 | 26.8 | 1.220 | 26.7 | 1.210 |
| OMe | 61.3 | 3.46 | 61.3 | 3.46 | |||||
| Compound | IC50 ± SE (µg/mL) a |
|---|---|
| holothurin A5 (1) | 1.9 ± 0.1 |
| holothurin A (2) | 1.4 ± 0.1 |
| echinoside A (3) | 1.2 ± 0.2 |
| 24-dehydroechinoside A (4) | 2.5 ± 0.4 |
| adriamycin | 0.6 ± 0.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grauso, L.; Yegdaneh, A.; Sharifi, M.; Mangoni, A.; Zolfaghari, B.; Lanzotti, V. Molecular Networking-Based Analysis of Cytotoxic Saponins from Sea Cucumber Holothuria atra. Mar. Drugs 2019, 17, 86. https://doi.org/10.3390/md17020086
Grauso L, Yegdaneh A, Sharifi M, Mangoni A, Zolfaghari B, Lanzotti V. Molecular Networking-Based Analysis of Cytotoxic Saponins from Sea Cucumber Holothuria atra. Marine Drugs. 2019; 17(2):86. https://doi.org/10.3390/md17020086
Chicago/Turabian StyleGrauso, Laura, Afsaneh Yegdaneh, Mohsen Sharifi, Alfonso Mangoni, Behzad Zolfaghari, and Virginia Lanzotti. 2019. "Molecular Networking-Based Analysis of Cytotoxic Saponins from Sea Cucumber Holothuria atra" Marine Drugs 17, no. 2: 86. https://doi.org/10.3390/md17020086