Understanding Forest Health with Remote Sensing, Part III: Requirements for a Scalable Multi-Source Forest Health Monitoring Network Based on Data Science Approaches
Abstract
:1. Introduction
- (1)
- What is required to bridge the gaps in information, data, models, and tools needed by forest managers and scientists for a better understanding of the complexity and multidimensionality of forest health drivers, stressors, disturbances, effects, and related processes?
- (2)
- Why are these requirements essential for a better understanding of forest health?
- (3)
- Which requirements are imperative to establish a multidimensional, multi-source forest health monitoring network in the future?
2. Forest Health
2.1. Understanding Forest Health
2.2. Indicator Requirements for Monitoring Forest Health
- They must reflect status, stress, disturbances, and resource limitations.
- They must be able to map different (i) organismic; (ii) temporal; and (iii) process characteristics, all across different scales.
- They should be recordable by different in situ monitoring approaches (forest inventory monitoring, biological and morphological species concepts, concept of ecological integrity, monitoring of animal distribution, anthropogenic drivers, or abiotic indicators (soil, water, climate, air), as well as close-range and air and spaceborne RS approaches on different platforms).
- It should be possible to standardize, objectively repeat, and thus compare indicators.
- Due to the heterogeneity and the complexity of different indicators in big data, all indicators should be transferable into digital form through a human–computer communication interpretation language to meet the requirements of data science.
2.3. Existing Standardized Approaches for Monitoring Forest Health
2.4. Initiatives to Unite Indicators, Data, and Approaches
- Set up a collection of harmonized Europe-wide information on the multi-functional role of forests and forest resources.
- Provision of harmonized forest datasets that were developed from EU funding, including Horizon2020, LIFE +, Copernicus, etc.
- Integration of diverse information systems (e.g., European Forest Fire Information System—EFFIS) and data platforms (e.g., European Forest Data Center, EFDAC) into a dynamic modular system that combines data (in situ forest data, forest data based on Copernicus RS information) and models into applications.
- Linking existing forest-related policy fields of the EU to relevant networks and initiatives in the context of forest information.
- Foster the availability of geo-data that are standardized throughout Europe, irrespective of national borders and with a high degree of spatial accuracy, in line with the INSPIRE directive (infrastructure for spatial information in Europe).
- Provision of a digital information on the distributions of the most important forest tree species and changes in forest dynamics.
- o
- Assessment of the suitability of tree species and forest types for current and future climate conditions, biophysical mapping, and assessment of ecosystem services in the forest;
- o
- Measurement of changes to the provision of these services as a consequence of anthropogenic and natural disturbances; economic assessment and ecological and economic audits.
- Several modules have been developed within FISE: (i) forests and natural disturbances such as fires and pests; (ii) forest and the bioeconomy; (iii) forests and climate change; and (iv) forest and ecosystem services.
3. Requirements to Understand the Effects of Multiple Stressors on Forest Health
4. Requirements for Using RS Approaches to Monitor FH
4.1. Requirements for Spatial Short and Long-Term Ground Truth Measurements on Different Scales
4.2. Requirements for Merging Different FH Modeling Approaches
4.3. Requirements for Linking Different Approaches to RS Techniques
4.4. Requirements for Merging RS Data from Different Sources
4.5. Requirements for Understanding and Handling RS Uncertainties
4.6. Requirements for Standardizing RS Monitoring Approaches
5. Requirements for Coupling Different Monitoring Approaches
5.1. In Situ Approaches beyond Forest Inventory Approaches
5.2. RS Approach for FH Monitoring
5.3. Phenotyping
5.4. Ecological Integrity
6. Requirements for Using Data Science as a Bridge
6.1. Open Science
6.1.1. Open Data Policy and Free Data Access
6.1.2. Open Science Clouds
6.2. Standardization in Data Analysis
6.2.1. Standardized Data Management Approaches
6.2.2. Standardization of Essential Variables of Forest Ecosystems
6.2.3. Standardized in Data Analysis (Thematic Exploitation Platforms)
6.3. Digitalization
6.3.1. Human-Driven Monitoring
6.3.2. Digitalization and Big Data
6.4. The Semantic Web
6.4.1. Semantification
6.4.2. Ontologization
6.4.3. Linked Open Data Approaches
6.5. Proof, Trust, and Uncertainty in Data Science
6.6. Data Science Analysis
6.6.1. Data Mining, Machine Learning, Artifizielle Intelligenz
6.6.2. Handling Petabyte-Scale RS Data with Cloud and Web Services
6.7. Easy-to-Handle Environmental Assessment and Decision-Making Support Systems
- Environmental assessment and decision-making must meet the requirements of data science. In particular, this means handling big data with high volume, velocity, variety, and veracity, and to integrate open data.
- Tools must enable extensive algorithms for data mining and machine learning.
- Systems must contain interfaces to the Semantic Web, thus enabling the analysis of semantically linked heterogeneous information via linked open data.
- Systems must allow for the assessment of uncertainties of in situ monitoring, RS data, and data science in the assessment and the resulting decision.
- Data science analyses should be possible with a simple coupling and management with other systems such as Haadop®, databanks, Web Services, Semantic Web Services, or cloud computing, as well as the use of the Google Earth Engine.
- Systems should be implementable on simple computers that are only available for authorities, users, and decision-makers.
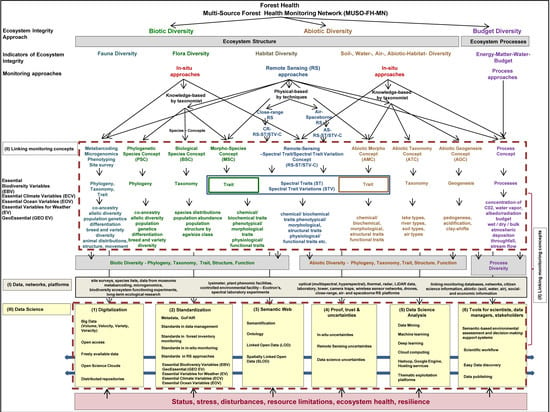
7. Requirements for a Future Multi-Source Forest Health Monitoring Network (MUSO-FH-MN)
- Digitalization: The digitalization of FH data and information will determine the receptiveness of the forest analysis process in the future MUSO-FH-MN. Crucial criteria and elements of the digitalization process are: not all elements can be digitally monitored in the digitalization process, and consequently, one must differentiate between human-driven monitoring and digital-driven monitoring elements. Furthermore, the following aspects are all important to foster the digitalization process: open access to tools, software, algorithms, instruments, or platforms, freely available data, free policy for species and RS and geodata [55], abiotic, socio-economic and other geo-data, the development and use of Open Science Clouds [175], Thematic Exploitation Platforms, the handling of big FH data with high volume, velocity, variety and veracity, as well as the management of distributed repositories.
- Standardization: FH data, information, indicators, data management, various FH monitoring approaches, tools, algorithms, and models all have to be standardized, administered, stored, processed, updated, as well as linked and evaluated with other platforms and networks. The basic standardization and the basic elements of data science are effective metadata management based on the principles of FAIR with the four criteria of Findability, Accessibility, Interoperability, and Reusability of metadata [191]. Standards in data management, standards in forest inventory monitoring, in situ monitoring beyond forest inventory monitoring for animals and plant species, as well as standards in RS approaches are crucial elements in data science. Furthermore, it is imperative to implement and integrate various existing concepts of essential variables such as the Essential Climate Variables (ECV) [194], the Essential Variables for Weather—EVW [195], the Essential Ocean Variables—EOV [193], the Essential Biodiversity Variables (EBV) [192], in order to develop the Essential GeoVariables—GEV GeoEssential (http://www.geoessential.net/), as well as develop the essential variables for domains such as agriculture, soils, catastrophes, ecosystems, health, and urban development.
- Semantic Web: The linking of complex, heterogeneous, and multidimensional FH information, indicators, data, Internet of FH Things (IOT), information, monitoring approaches, tools, different scales, RS platforms, and models as well as assessment and decision-making support systems for scientists, data managers, and stakeholders in a semantic-enabling way according to the standards of the World Wide Web Consortium [205] is an important step to cope with the human–computer communication process and couple complex FH data, information, models and platforms. Important elements of the semantic web are: semantification, ontologization [206], Linked Open Data approaches [248] and Spatially Linked Open Data approaches [189].
- Proof, Trust, and Uncertainties: Unlike many other research areas, most approaches, data, information or models used in data science involve a certain level of uncertainty i.e., uncertainties in forestry inventory monitoring information, in situ uncertainties, RS uncertainties, and data science uncertainties.
- Data Science Analysis: The digitalization of the world and forest ecosystem components requires handling Big Data along with its four aspects: volume, velocity, variety, and veracity. Consequently, data science analysis requires methods of data mining, machine learning, deep learning, tools, systems, or platforms such as Hadoop, Google Engine, Hosting services, Semantic Web Services, cloud computing, as well as Thematic Exploitation Platforms. Here, deep learning is central to identifying, processing, and analyzing patterns in automation and new innovations of FH analysis and assessment.
- Tools for scientists, data managers, and stakeholders: Crucial elements for a data-driven, fast, objective, applicable, and implementable decision-making support system for forest managers, stakeholders, and politicians are: open, easy handling and data science-based environmental assessment and decision-making support systems, comprehendible and easy-to-operate scientific workflows, and easy and up-to-date data publishing tools.
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lausch, A.; Erasmi, S.; King, D.J.; Magdon, P.; Heurich, M. Understanding forest health by remote sensing—Part II—A review of approaches and data models. Remote Sens. 2017, 9, 129. [Google Scholar] [CrossRef]
- Trumbore, S.; Brando, P.; Hartmann, H. Forest health and global change. Science 2015, 349, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Guerra, C.; Navarro, L.M.; Kissling, W.D.; London, M.C.; Turak, E.; Balvanera, P.; Costello, M.J.; Delavaud, A.; El Serafy, G.Y.; Ferrier, S.; et al. Monitoring biodiversity change through effective global coordination. Curr. Opin. Environ. Sustain. 2017, 29, 158–169. [Google Scholar]
- Krug, C.B.; Schaepman, M.E.; Shannon, L.J.; Cavender-bares, J.; Cheung, W.; Mcintyre, P.B.; Metzger, J.P.; Obura, D.O.; Schmid, B.; Strassburg, B.B.N.; et al. Observations, indicators and scenarios of biodiversity and ecosystem services change—A framework to support policy and decision-making. Curr. Opin. Environ. Sustain. 2017, 29, 198–206. [Google Scholar] [CrossRef]
- Tomppo, E.O.; Schadauer, K. Harmonization of National Forest Inventories in Europe: Advances under COST Action E43. For. Sci. 2012, 58, 191–200. [Google Scholar] [CrossRef]
- Tomppo, E.; Gschwantner, T.; Lawrence, M.; McRoberts, E.R. National Forest Inventories. Pathways for Common Reporting; Springer: Heidelberg, Germany; Dordrecht, The Netherlands; London, UK; New York, NY, USA, 2010. [Google Scholar]
- McRoberts, R.E.; Tomppo, E.O.; Schadauer, K.; Ståhl, G. Harmonizing national forest inventories. For. Sci. 2012, 58, 189–190. [Google Scholar] [CrossRef]
- Traub, B.; Meile, R.; Speich, S.; Rösler, E. The data storage and analysis system of the Swiss National Forest Inventory. Comput. Electron. Agric. 2017, 132, 97–107. [Google Scholar] [CrossRef]
- Henttonen, H.M.; Nöjd, P.; Mäkinen, H. Forest Ecology and Management Environment-induced growth changes in the Finnish forests during 1971–2010—An analysis based on National Forest Inventory. For. Ecol. Manag. 2017, 386, 22–36. [Google Scholar] [CrossRef]
- Bruelheide, H.; Nadrowski, K.; Assmann, T.; Bauhus, J.; Both, S.; Buscot, F.; Chen, X.Y.; Ding, B.; Durka, W.; Erfmeier, A.; et al. Designing forest biodiversity experiments: General considerations illustrated by a new large experiment in subtropical China. Methods Ecol. Evol. 2014, 5, 74–89. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 8992, 1–23. [Google Scholar] [CrossRef]
- Wulder, M.A.; Masek, J.G.; Cohen, W.B.; Loveland, T.R.; Woodcock, C.E. Opening the archive: How free data has enabled the science and monitoring promise of Landsat. Remote Sens. Environ. 2012, 122, 2–10. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Loveland, T.R.; Woodcock, C.E.; Belward, A.S.; Cohen, W.B.; Fosnight, E.A.; Shaw, J.; Masek, J.G.; Roy, D.P. The global Landsat archive: Status, consolidation, and direction. Remote Sens. Environ. 2016, 185, 271–283. [Google Scholar] [CrossRef]
- Turner, W.; Rondinini, C.; Pettorelli, N.; Mora, B.; Leidner, A.K.; Szantoi, Z.; Buchanan, G.; Dech, S.; Dwyer, J.; Herold, M.; et al. Free and open-access satellite data are key to biodiversity conservation. Biol. Conserv. 2015, 182, 173–176. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Tomppo, E.O. Remote sensing support for national forest inventories. Remote Sens. Environ. 2007, 110, 412–419. [Google Scholar] [CrossRef]
- Massey, A.; Mandallaz, D.; Lanz, A. Integrating remote sensing and past inventory data under the new annual design of the Swiss National Forest Inventory using three-phase design-based regression estimation. Can. J. For. Res. 2014, 44, 1177–1186. [Google Scholar] [CrossRef]
- Immitzer, M.; Stepper, C.; Böck, S.; Straub, C.; Atzberger, C. Use of WorldView-2 stereo imagery and National Forest Inventory data for wall-to-wall mapping of growing stock. For. Ecol. Manag. 2016, 359, 232–246. [Google Scholar] [CrossRef]
- Tomppo, E.; Olsson, H.; Ståhl, G.; Nilsson, M.; Hagner, O.; Katila, M. Combining national forest inventory field plots and remote sensing data for forest databases. Remote Sens. Environ. 2008, 112, 1982–1999. [Google Scholar] [CrossRef]
- Abegg, M.; Kükenbrink, D.; Zell, J.; Schaepman, M.E.; Morsdorf, F. Terrestrial Laser Scanning for Forest Inventories—Tree Diameter Distribution and Scanner Location Impact on Occlusion. Forests 2017, 8, 184. [Google Scholar] [CrossRef]
- Barrett, F.; McRoberts, R.E.; Tomppo, E.; Cienciala, E.; Waser, L.T. A questionnaire-based review of the operational use of remotely sensed data by national forest inventories. Remote Sens. Environ. 2016, 174, 279–289. [Google Scholar] [CrossRef]
- FAO. State of Europe’s Forests 2015. Ministerial Conference on the Protection of Forests in Europe FOREST EUROPE; Liaison Unit Madrid: Madrid, Spain, 2015. [Google Scholar]
- Asner, G.P.; Anderson, C.B.; Martin, R.E.; Tupayachi, R.; Knapp, D.E.; Sinca, F. Landscape biogeochemistry reflected in shifting distributions of chemical traits in the Amazon forest canopy. Nat. Geosci. 2015, 8, 567–573. [Google Scholar] [CrossRef]
- Buermann, W.; Parida, B.; Jung, M.; MacDonald, G.M.; Tucker, C.J.; Reichstein, M. Recent shift in Eurasian boreal forest greening response may be associated with warmer and drier summers. Geophys. Res. Lett. 2014, 41, 1995–2002. [Google Scholar] [CrossRef]
- Lausch, A.; Erasmi, S.; King, D.J.; Magdon, P.; Heurich, M. Understanding Forest Health with Remote Sensing—Part I—A Review of Spectral Traits, Processes and Remote-Sensing Characteristics. Remote Sens. 2016, 8, 1029. [Google Scholar] [CrossRef]
- Lausch, A.; Bannehr, L.; Beckmann, M.; Boehm, C.; Feilhauer, H.; Hacker, J.M.; Heurich, M.; Jung, A.; Klenke, R.; Neumann, C.; et al. Linking Earth Observation and taxonomic, structural and functional biodiversity: Local to ecosystem perspectives. Ecol. Indic. 2016, 70, 317–339. [Google Scholar] [CrossRef]
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; Mace, G.M.; Tilman, D.; Wardle, D.A.; et al. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Galic, N.; Sullivan, L.L.; Grimm, V.; Forbes, V.E. When things don’t add up: Quantifying impacts of multiple stressors from individual metabolism to ecosystem processing. Ecol. Lett. 2018, 21, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Lausch, A.; Blaschke, T.; Haase, D.; Herzog, F.; Syrbe, R.-U.; Tischendorf, L.; Walz, U. Understanding and quantifying landscape structure—A review on relevant process characteristics, data models and landscape metrics. Ecol. Model. 2015, 295, 31–41. [Google Scholar] [CrossRef]
- Schmidt, J.; Fassnacht, F.E.; Lausch, A.; Schmidtlein, S. Assessing the functional signature of heathland landscapes via hyperspectral remote sensing. Ecol. Indic. 2017, 73, 505–512. [Google Scholar] [CrossRef]
- Schmidtlein, S.; Feilhauer, H.; Bruelheide, H. Mapping plant strategy types using remote sensing. J. Veg. Sci. 2012, 23, 395–405. [Google Scholar] [CrossRef]
- Rapport, D.J.; Costanza, R.; McMichael, A.J. Assessing ecosystem health. Trends Ecol. Evol. 1998, 13, 397–402. [Google Scholar] [CrossRef]
- Müller, F. Indicating ecosystem and landscape organisation. Ecol. Indic. 2005, 5, 280–294. [Google Scholar] [CrossRef]
- Haase, P.; Tonkin, J.D.; Stoll, S.; Burkhard, B.; Frenzel, M.; Geijzendorffer, I.R.; Häuser, C.; Klotz, S.; Kühn, I.; McDowell, W.H.; et al. The next generation of site-based long-term ecological monitoring: Linking essential biodiversity variables and ecosystem integrity. Sci. Total Environ. 2018, 613, 1376–1384. [Google Scholar] [CrossRef] [PubMed]
- Lausch, A.; Olaf, B.; Stefan, K.; Leitao, P.; Jung, A.; Rocchini, D.; Schaepman, M.E.; Skidmore, A.K.; Tischendorf, L.; Knapp, S. Understanding and assessing vegetation health by in-situ species and remote sensing approaches Journal. Methods Ecol. Evol. 2018. [Google Scholar] [CrossRef]
- Johann Heinrich von Thünen-Institute, Forest Condition Monitoring (FCM) Level-I-Monitoring. Available online: https://www.thuenen.de/de/wo/arbeitsbereiche/waldmonitoring/ (accessed on 3 January 2017).
- Lorenz, M. International co-operative programme on assessment and monitoring of air pollution effects on forests—ICP Forests. Water Air Soil Pollut. 1995, 85, 1221–1226. [Google Scholar] [CrossRef]
- Federal Ministry for Food and Agriculture, National Forest Inventory Level-III-Monitoring. Available online: https://bwi.info/ (accessed on 3 January 2017).
- United States Department of Agriculture (USDA) Forest Service, FH Monitoring (M). Available online: https://www.fs.fed.us/foresthealth/monitoring/index.shtml (accessed on 3 January 2017).
- Yang, J.; Dai, G.; Wang, S. China’s national monitoring program on ecological functions of forests: An analysis of the protocol and initial results. Forests 2015, 6, 809–826. [Google Scholar] [CrossRef]
- Canadian Forest Service (CFS), National FHM Network. Available online: http://www.cfs.nrcan.gc.ca/publications/?id=4105 (accessed on 3 January 2017).
- National Forest Inventory (NFI), Canada, National Forest Inventory. Available online: https://nfi.nfis.org/en/ (accessed on 3 January 2017).
- United Nations Economic Commission for Europe (UNECE), ICP. Available online: http://icp-forests.net/, http://icp-forests.net/page/icp-forests-executive-report (accessed on 3 January 2017).
- Food and Agriculture Organization of the United Nations (FAO), Forest Resources Assessment. Available online: http://www.fao.org/forest-resources-assessment/en/ (accessed on 3 January 2017).
- Kandler, G. The design of the second German national forest inventory. In Proceedings of the Eighth Annual Forest Inventory and Analysis Symposium; McRoberts, R.E., Reams, G.A., Van Deusen, P.C., McWilliams, W.H., Eds.; Monterey, CA. Gen. Tech. Report WO-79; U.S. Department of Agriculture, Forest Service: Washington, DC, USA, 2009; pp. 19–24. [Google Scholar]
- Matyssek, R.; Wieser, G.; Calfapietra, C.; de Vries, W.; Dizengremel, P.; Ernst, D.; Jolivet, Y.; Mikkelsen, T.N.; Mohren, G.M.J.; Le Thiec, D.; et al. Forests under climate change and air pollution: Gaps in understanding and future directions for research. Environ. Pollut. 2012, 160, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Pause, M.; Schweitzer, C.; Rosenthal, M.; Keuck, V.; Bumberger, J.; Dietrich, P.; Heurich, M.; Jung, A.; Lausch, A. In situ/remote sensing integration to assess forest health—A review. Remote Sens. 2016, 8, 471. [Google Scholar] [CrossRef]
- Turner, W.; Spector, S.; Gardiner, N.; Fladeland, M.; Sterling, E.; Steininger, M. Remote sensing for biodiversity science and conservation. Trends Ecol. Evol. 2003, 18, 306–314. [Google Scholar] [CrossRef]
- Toth, C.; Jóźków, G. Remote sensing platforms and sensors: A survey. ISPRS J. Photogramm. Remote Sens. 2016, 115, 22–36. [Google Scholar] [CrossRef]
- Kuenzer, C.; Blümel, A.; Gebhardt, S.; Quoc, T.V.; Dech, S. Remote sensing of mangrove ecosystems: A review. Remote Sens. 2011, 3, 878–928. [Google Scholar] [CrossRef]
- Banskota, A.; Kayastha, N.; Falkowski, M.J.; Wulder, M.A.; Froese, R.E.; White, J.C. Forest monitoring using Landsat time series data: A review. Can. J. Remote Sens. 2014, 40, 362–384. [Google Scholar] [CrossRef]
- Joshi, N.; Mitchard, E.T.A.; Woo, N.; Torres, J.; Moll-rocek, J.; Ehammer, A. Mapping dynamics of deforestation and forest degradation in tropical forests using radar satellite data. Environ. Res. Lett. 2015, 10, 34014. [Google Scholar] [CrossRef]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Kuenzer, C.; Ottinger, M.; Wegmann, M.; Guo, H. Earth observation satellite sensors for biodiversity monitoring: Potentials and bottlenecks. Int. J. Remote Sens. 2014, 35, 6599–6647. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Anderson, C.B.; Martin, R.E.; Vaughn, N. Large-scale climatic and geophysical controls on the leaf economics spectrum. Proc. Natl. Acad. Sci. USA 2016, 113, E4043–E4051. [Google Scholar] [CrossRef] [PubMed]
- Wulder, M.A.; Coops, N.C. Make Earth observations open access. Nature 2014, 513, 30–31. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, M.J.; Brockerhoff, E.G.; Wingfield, B.D.; Slippers, B. Planted forest health: The need for a global strategy. Science 2015, 349, 832–836. [Google Scholar] [CrossRef] [PubMed]
- McDowell, N.G.; Coops, N.C.; Beck, P.S.A.; Chambers, J.Q.; Gangodagamage, C.; Hicke, J.A.; Huang, C.; Kennedy, R.; Krofcheck, D.J.; Litvak, M.; et al. Global satellite monitoring of climate-induced vegetation disturbances. Trends Plant Sci. 2015, 20, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Jetz, W.; Cavender-Bares, J.; Pavlick, R.; Schimel, D.; Davis, F.W.; Asner, G.P.; Guralnick, R.; Kattge, J.; Latimer, A.M.; Moorcroft, P.; et al. Monitoring plant functional diversity from space. Nat. Plants 2016, 2, 1–5. [Google Scholar]
- Riemann, R.; Wilson, B.T.; Lister, A.; Parks, S. An effective assessment protocol for continuous geospatial datasets of forest characteristics using USFS Forest Inventory and Analysis (FIA) data. Remote Sens. Environ. 2010, 114, 2337–2352. [Google Scholar] [CrossRef]
- Sader, S.A.; Hoppus, M.; Metzler, J.W.; Jin, S. Perspectives of Maine forest cover change from Landsat imagery and Forest Inventory Analysis (FIA). J. For. 2005, 103, 299–303. [Google Scholar]
- Verheyen, K.; Vanhellemont, M.; Auge, H.; Baeten, L.; Baraloto, C.; Barsoum, N.; Bilodeau-Gauthier, S.; Bruelheide, H.; Castagneyrol, B.; Godbold, D.; et al. Contributions of a global network of tree diversity experiments to sustainable forest plantations. Ambio 2016, 45, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Verhaeghe, G. Trees4 future-Designing Trees for the Future. Access to Key Research Infrastructures and Tools in the Field of Forestry and Wood Technology in Europe. Drewno 2012, 55, 141–144. [Google Scholar]
- Baeten, L.; Verheyen, K.; Wirth, C.; Bruelheide, H.; Bussotti, F.; Finér, L.; Jaroszewicz, B.; Selvi, F.; Valladares, F.; Allan, E.; et al. A novel comparative research platform designed to determine the functional significance of tree species diversity in European forests. Perspect. Plant Ecol. Evol. Syst. 2013, 15, 281–291. [Google Scholar] [CrossRef]
- Scherer-Lorenzen, M.; Schulze, E.D.; Don, A.; Schumacher, J.; Weller, E. Exploring the functional significance of forest diversity: A new long-term experiment with temperate tree species (BIOTREE). Perspect. Plant Ecol. Evol. Syst. 2007, 9, 53–70. [Google Scholar] [CrossRef]
- Watch, G.F. Global Forest Watch. For. Chron. 2000, 76, 398. [Google Scholar]
- Showstack, R. Global Forest Watch Initiative Provides Opportunity for Worldwide Monitoring. EOS Trans. Am. Geophys. Union 2014, 95, 77–79. [Google Scholar] [CrossRef]
- Gracan, J.; Kajba, D.; Peric, S.; Idzojtic, M.; Ivankovic, M.; Marjanovic, H. EUFORGEN forest genetic resources programme fourth EUFORGEN: Steering committee meeting. Progr. Eur. Sumski Genet. Resur. (EUFORGEN) 2004, 128, 689–697. [Google Scholar]
- Milly, P.C.D. Climate, soil water storage, and the average water balance. Water Resour. Res. 1994, 30, 2143–2156. [Google Scholar] [CrossRef]
- Struthers, I.; Hinz, C.; Sivapalan, M.; Deutschmann, G.; Beese, F.; Meissner, R. Modelling the water balance of a free-draining lysimeter using the downward approach. Hydrol. Process. 2003, 17, 2151–2169. [Google Scholar] [CrossRef]
- Will, G.M. A field lysimeter to study water movement and nutrient content in a pumice soil under Pinus radiata forest. I. Site and construction details. N. Z. J. For. Sci. 1977, 7, 144–150. [Google Scholar]
- Müller, J. Forestry and water budget of the lowlands in northeast Germany—Consequences for the choice of tree species and for forest management. J. Water Land Dev. 2009, 13A, 133–148. [Google Scholar] [CrossRef]
- Johnson, R.; Williams, L.; Ayars, J.; Trout, T. Weighing lysimeters aid study of water relations in tree and vine crops. Calif. Agric. 2005, 59, 133–136. [Google Scholar] [CrossRef]
- Ben-Gal, A.; Kool, D.; Agam, N.; van Halsema, G.E.; Yermiyahu, U.; Yafe, A.; Presnov, E.; Erel, R.; Majdop, A.; Zipori, I.; et al. Whole-tree water balance and indicators for short-term drought stress in non-bearing ‘Barnea’ olives. Agric. Water Manag. 2010, 98, 124–133. [Google Scholar] [CrossRef]
- Yick, J.; Mukherjee, B.; Ghosal, D. Wireless sensor network survey. Comput. Netw. 2008, 52, 2292–2330. [Google Scholar] [CrossRef]
- Marchetti, M. Monitoring and Indicators of Forest Biodiversity in Europe: From Ideas to Operationality; European Forest Institute: Joensuu, Finland, 2005; Volume 58. [Google Scholar]
- Großkinsky, D.K.; Svensgaard, J.; Christensen, S.R.T. Plant phenomics and the need for physiological phenotyping across scales to narrow the genotype-to-phenotype knowledge gap. J. Exp. Bot. 2015, 66, 5429–5440. [Google Scholar] [CrossRef] [PubMed]
- Pieruschka, R.; Lawson, T. Preface. J. Exp. Bot. 2015, 66, 5385–5387. [Google Scholar] [CrossRef] [PubMed]
- Furbank, R.T. Foreword: Plant phenomics: From gene to form and function. Funct. Plant Biol. 2009, 36, 5–6. [Google Scholar]
- Großkinsky, D.K.; Pieruschka, R.; Svensgaard, J.; Rascher, U.; Christensen, S.; Schurr, U.; Roitsch, T. Phenotyping in the fields: Dissecting the genetics of quantitative traits and digital farming. New Phytol. 2015, 207, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Blumwald, E. Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 2010, 61, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Lawton, J.H.; Naeem, S.; Woodfin, R.M.; Brown, V.K.; Gange, A.; Godfray, H.C.J.; Heads, P.A.; Lawler, S.; Magda, D.; Thomas, C.D.; et al. The Ecotron: A controlled environmental facility for the investigation of population and ecosystem processes. Philos. Trans. Biol. Sci. 1993, 341, 181–194. [Google Scholar] [CrossRef]
- Ehrhardt, D.W.; Frommer, W.B. New Technologies for 21st Century Plant Science. Plant Cell 2012, 24, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [PubMed]
- Kuijken, R.C.P.; Van Eeuwijk, F.A.; Marcelis, L.F.M.; Bouwmeester, H.J. Root phenotyping: From component trait in the lab to breeding. J. Exp. Bot. 2015, 66, 5389–5401. [Google Scholar] [CrossRef] [PubMed]
- Wardle, D.A. Do experiments exploring plant diversity-ecosystem functioning relationships inform how biodiversity loss impacts natural ecosystems? J. Veg. Sci. 2016, 27, 646–653. [Google Scholar] [CrossRef]
- Loreau, M. Linking biodiversity and ecosystems: Towards a unifying ecological theory. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, N.; Barnes, A.D.; Cesarz, S.; Craven, D.; Ferlian, O.; Gottschall, F.; Hines, J.; Sendek, A.; Siebert, J.; Thakur, M.P.; et al. Biodiversity-ecosystem function experiments reveal the mechanisms underlying the consequences of biodiversity change in real world ecosystems. J. Veg. Sci. 2016, 27, 1061–1070. [Google Scholar] [CrossRef]
- Purvis, A.; Hector, A. Getting the measure of biodiversity. Nature 2000, 405, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Gaston, K.J.; Chown, S.L.; Calosi, P.; Bernardo, J.; Bilton, D.T.; Clarke, A.; Clusella-Trullas, S.; Ghalambor, C.K.; Konarzewski, M.; Peck, L.S.; et al. Macrophysiology: A Conceptual Reunification. Am. Nat. 2009, 174, 595–612. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Kryston, K.; Vaughn, N.; Knapp, D.E.; Bentley, L.P.; Shenkin, A.; Salinas, N.; Sinca, F.; et al. Scale dependence of canopy trait distributions along a tropical forest elevation gradient. New Phytol. 2016, 214, 937–988. [Google Scholar] [CrossRef] [PubMed]
- Buddenbaum, H.; Stern, O.; Paschmionka, B.; Hass, E.; Gattung, T.; Stoffels, J.; Hill, J.; Werner, W. Using VNIR and SWIR field imaging spectroscopy for drought stress monitoring of beech seedlings. Int. J. Remote Sens. 2015, 36, 4590–4605. [Google Scholar] [CrossRef]
- Brosinsky, A.; Lausch, A.; Doktor, D.; Salbach, C.; Merbach, I.; Gwillym-Margianto, S.; Pause, M. Analysis of Spectral Vegetation Signal Characteristics as a Function of Soil Moisture Conditions Using Hyperspectral Remote Sensing. J. Indian Soc. Remote Sens. 2013, 42, 311–324. [Google Scholar] [CrossRef]
- Zacharias, S.; Bogena, H.; Samaniego, L.; Mauder, M.; Fuß, R.; Pütz, T.; Frenzel, M.; Schwank, M.; Baessler, C.; Butterbach-Bahl, K.; et al. A Network of Terrestrial Environmental Observatories in Germany. Vadose Z. J. 2011, 10, 955. [Google Scholar] [CrossRef]
- Borg, E.; Schiller, C.; Daedelow, H.; Fichtelmann, B.; Jahncke, D.; Renke, F.; Tamm, H.; Asche, H. Automated Generation of Value-Added Products for the Validation of Remote Sensing Information Based on In-situ Data. In Proceedings of the International Conference on Computational Science and Its Applications, Guimarães, Portugal, 30 June–3 July 2014; pp. 393–407. [Google Scholar]
- Jülich, F.; Centre, H. TERENO: German network of terrestrial environmental observatories. J. Large Scale Res. Facil. 2016, 52, 1–8. [Google Scholar]
- Wollschläger, U.; Attinger, S.; Borchardt, D.; Brauns, M.; Cuntz, M.; Dietrich, P.; Fleckenstein, J.H.; Friese, K.; Friesen, J.; Harpke, A.; et al. The Bode Hydrological Observatory: A platform for integrated, interdisciplinary hydro-ecological research within the TERENO Harz/Central German Lowland Observatorytle. Environ. Earth Sci. 2010, 76, 29. [Google Scholar] [CrossRef]
- Vihervaara, P.; D’Amato, D.; Forsius, M.; Angelstam, P.; Baessler, C.; Balvanera, P.; Boldgiv, B.; Bourgeron, P.; Dick, J.; Kanka, R.; et al. Using long-term ecosystem service and biodiversity data to study the impacts and adaptation options in response to climate change: Insights from the global ILTER sites network. Curr. Opin. Environ. Sustain. 2013, 5, 53–66. [Google Scholar] [CrossRef]
- Müller, F.; Baessler, C.; Schubert, H.; Klotz, S. (Eds.) Long-Term Ecological Research. Between Theory and Application; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Riddering, J.; Holden, Z.A.; Jolly, W.M.; Warren, A. Smartphone applications for data collections, dynamic modeling, and visualization in the wildland fire environment. Fire Manag. Today 2015, 74, 10–15. [Google Scholar]
- Aanensen, D.M.; Huntley, D.M.; Feil, E.J.; Al-Own, F.; Spratt, B.G. EpiCollect: Linking smartphones to web applications for epidemiology, ecology and community data collection. PLoS ONE 2009, 4, e6968. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.; Sullivan, B.; Iliff, M.; Fink, D.; Kelling, S. eBird: Engaging birders in science and conservation. PLoS Biol. 2011, 9, e1001220. [Google Scholar] [CrossRef] [PubMed]
- Priess, J.A.; Elger, R.H.J. The ESM-App—A new smartphone application to map ecosystem services. In Proceedings of the 7th International Congress on Environmental Modelling and Software, San Diego, CA, USA, 15–19 June 2014; Ames, D.P., Quinn, N.W.T., Rizzoli, A.E., Eds.; International Environmental Modelling and Software Society: Leipzig, Germany, 2014; ISBN 978-88-9035-744-2. [Google Scholar]
- U.S. Department of Commerce, N.O. and A.A. The mPING Project. 2016. Available online: http://www.nssl.noaa.gov/projects/ping (accessed on 31 December 2016).
- Eberle, J.; Hüttich, C.; Schmulius, C. MySeason. App from Institute for Geography, Department for Earth Observation, 2016. Available online: http://www.eo.uni-jena.de (accessed on 31 December 2016).
- Borg, E. CAL/VAL Site DEMMIN for Remote Sensing; Network of European Regions Using Space Technology: Brussels, Belgium, 2010. [Google Scholar]
- Shugart, H.H.; Asner, G.P.; Fischer, R.; Huth, A.; Knapp, N.; Le Toan, T.; Shuman, J.K. Computer and remote-sensing infrastructure to enhance large-scale testing of individual-based forest models. Front. Ecol. Environ. 2015, 13, 503–511. [Google Scholar] [CrossRef]
- Mäkinen, A.; Holopainen, M.; Kangas, A.; Rasinmäki, J. Propagating the errors of initial forest variables through stand- and tree-level growth simulators. Eur. J. For. Res. 2010, 129, 887–897. [Google Scholar] [CrossRef]
- Fischer, R.; Bohn, F.; Dantas de Paula, M.; Dislich, C.; Groeneveld, J.; Gutiérrez, A.G.; Kazmierczak, M.; Knapp, N.; Lehmann, S.; Paulick, S.; et al. Lessons learned from applying a forest gap model to understand ecosystem and carbon dynamics of complex tropical forests. Ecol. Model. 2016, 326, 124–133. [Google Scholar] [CrossRef]
- Coops, N.C.; Waring, R.H.; Brown, S.R.; Running, S.W. Comparisons of predictions of net primary production and seasonal patterns in water use derived with two forest growth model in Southwestern Oregon. Ecol. Model. 2001, 142, 61–81. [Google Scholar] [CrossRef]
- Fawcett, D.; Verhoef, W.; Schläpfer, D.; Schneider, F.D.; Schaepman, M.E.; Damm, A. Remote Sensing of Environment Advancing retrievals of surface reflectance and vegetation indices over forest ecosystems by combining imaging spectroscopy, digital object models, and 3D canopy modelling. Remote Sens. Environ. 2018, 204, 583–595. [Google Scholar] [CrossRef]
- Buddenbaum, H.; Hill, J. PROSPECT Inversions of Leaf Laboratory Imaging Spectroscopy—A Comparison of Spectral Range and Inversion Technique Influences. Photogramm. Fernerkundung Geoinf. 2015, 2015, 231–240. [Google Scholar] [CrossRef]
- Roedig, E.; Matthias, C.; Heinke, J.; Rammig, A.; Huth, A. Spatial heterogeneity of biomass and forest structure of the Amazon rainforest: Linking remote sensing, forest modeling and field inventory. Glob. Ecol. Biogeogr. 2017, 26, 1292–1302. [Google Scholar] [CrossRef]
- Pütz, S.; Groeneveld, J.; Henle, K.; Knogge, C.; Martensen, A.C.; Metz, M.; Metzger, J.P.; Ribeiro, M.C.; de Paula, M.D.; Huth, A. Long-term carbon loss in fragmented Neotropical forests. Nat. Commun. 2014, 5, 5037. [Google Scholar] [CrossRef] [PubMed]
- Brinck, K.; Fischer, R.; Groeneveld, J.; Lehmann, S.; Dantas De Paula, M.; Pütz, S.; Sexton, J.O.; Song, D.; Huth, A. High resolution analysis of tropical forest fragmentation and its impact on the global carbon cycle. Nat. Commun. 2017, 8, 14855. [Google Scholar] [CrossRef] [PubMed]
- Cernansky, R. Biodiversity moves beyond counting species. Nature 2017, 546, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Abelleira Martínez, O.J.; Fremier, A.K.; Günter, S.; Ramos Bendaña, Z.; Vierling, L.; Galbraith, S.M.; Bosque-Pérez, N.A.; Ordoñez, J.C. Scaling up functional traits for ecosystem services with remote sensing: Concepts and methods. Ecol. Evol. 2016, 6, 4359–4371. [Google Scholar] [CrossRef] [PubMed]
- Gamba, P.; Chanussot, J. Foreword to the Special Issue on Data Fusion. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1283–1288. [Google Scholar] [CrossRef]
- Ghassemian, H. A review of remote sensing image fusion methods. Inf. Fusion 2016, 32, 75–89. [Google Scholar] [CrossRef]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Linke, J.; McDermid, G.; Masek, J.G.; Gao, F.; White, J.C. A new data fusion model for high spatial- and temporal-resolution mapping of forest disturbance based on Landsat and MODIS. Remote Sens. Environ. 2009, 113, 1613–1627. [Google Scholar] [CrossRef]
- Gao, F.; Masek, J.; Schwaller, M.; Hall, F. On the blending of the landsat and MODIS surface reflectance: Predicting daily landsat surface reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2207–2218. [Google Scholar]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Seitz, N.; White, J.C.; Gao, F.; Masek, J.G.; Stenhouse, G. Generation of dense time series synthetic Landsat data through data blending with MODIS using a spatial and temporal adaptive reflectance fusion model. Remote Sens. Environ. 2009, 113, 1988–1999. [Google Scholar] [CrossRef]
- Torabzadeh, H.; Morsdorf, F.; Schaepman, M.E. Fusion of imaging spectroscopy and airborne laser scanning data for characterization of forest ecosystems—A review. ISPRS J. Photogramm. Remote Sens. 2014, 97, 25–35. [Google Scholar] [CrossRef]
- Taylor, J. Introduction to Error Analysis, the Study of Uncertainties in Physical Measurements, 2nd ed.; University Science Books: New York, NY, USA, 1997. [Google Scholar]
- Der Kiureghian, A.; Ditlevsen, O. Aleatory or epistemic? Does it matter? Struct. Saf. 2009, 31, 105–112. [Google Scholar] [CrossRef]
- Lane, D.A.; Maxfield, R.R. Ontological uncertainty and innovation. J. Evol. Econ. 2005, 15, 3–50. [Google Scholar] [CrossRef]
- Wu, H.; Li, Z.L. Scale issues in remote sensing: A review on analysis, processing and modeling. Sensors 2009, 9, 1768–1793. [Google Scholar] [CrossRef] [PubMed]
- Segl, K.; Guanter, L.; Rogass, C.; Kuester, T.; Roessner, S.; Kaufmann, H.; Sang, B.; Mogulsky, V.; Hofer, S. EeteSThe EnMAP end-to-end simulation tool. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 522–530. [Google Scholar] [CrossRef]
- Jung, A.; Götze, C.; Glässer, C. Overview of Experimental Setups in Spectroscopic Laboratory Measurements—The SpecTour Project. Photogramm. Fernerkundung Geoinf. 2012, 4, 433–442. [Google Scholar] [CrossRef]
- Baldocchi, D.; Falge, E.; Gu, L.H.; Olson, R.; Hollinger, D.; Running, S.; Anthoni, P.; Bernhofer, C.; Davis, K.; Evans, R.; et al. FLUXNET: A New Tool to Study the Temporal and Spatial Variability of Ecosystem-Scale Carbon Dioxide, Water Vapor, and Energy Flux Densities. Bull. Am. Meteorol. Soc. 2001, 82, 2415–2434. [Google Scholar] [CrossRef]
- Hueni, A.; Nieke, J.; Schopfer, J.; Kneubühler, M.; Itten, K.I. The spectral database SPECCHIO for improved long-term usability and data sharing. Comput. Geosci. 2009, 35, 557–565. [Google Scholar] [CrossRef]
- Saarinen, N.; White, J.C.; Wulder, M.A.; Kangas, A.; Tuominen, S.; Kankare, V.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Landsat archive holdings for Finland: Opportunities for forest monitoring. Forests 2018, 52, 9986. [Google Scholar] [CrossRef]
- Eldredge, N.; Cracraft, J. Phylogenetic Patterns and the Evolutionary Process; University Press: New York, NY, USA, 1980. [Google Scholar]
- Mayr, E. Systematics and the Origin of Species from the Viewpoint of a Zoologist; Columbia University Press: New York, NY, USA, 1942. [Google Scholar]
- Boulangeat, I.; Thuiller, W. Accounting for dispersal and biotic interactions to disentangle the drivers of species distributions and their abundances. Ecol. Lett. 2012, 15, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Violle, C.; Reich, P.B.; Pacala, S.W.; Enquist, B.J.; Kattge, J. The emergence and promise of functional biogeography. Proc. Natl. Acad. Sci. USA 2014, 111, 13690–13696. [Google Scholar] [CrossRef] [PubMed]
- Mayr, E.; Ashlock, P.D. Principles of Systematic Zoology; Systematic Biology: Oxford, UK, 1969. [Google Scholar]
- Green, J.L.; Bohannan, J.M.; Whitaker, R.J. Microbial biogeography: From taxonomy to traits. Science 2008, 320, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Knapp, S.; Kühn, I.; Wittig, R.; Ozinga, W.A.; Poschlod, P.; Klotz, S. Urbanization causes shifts in species’ trait state frequencies. Preslia 2008, 80, 375–388. [Google Scholar]
- Garnier, E.; Lavorel, S.; Ansquer, P.; Castro, H.; Cruz, P.; Dolezal, J.; Eriksson, O.; Fortunel, C.; Freitas, H.; Golodets, C.; et al. Assessing the effects of land-use change on plant traits, communities and ecosystem functioning in grasslands: A standardized methodology and lessons from an application to 11 European sites. Ann. Bot. 2007, 99, 967–985. [Google Scholar] [CrossRef] [PubMed]
- Knapp, S.; Kühn, I.; Stolle, J.; Klotz, S. Changes in the functional composition of a Central European urban flora over three centuries. Perspect. Plant Ecol. Evol. Syst. 2010, 12, 235–244. [Google Scholar] [CrossRef]
- Gámez-Virués, S.; Perović, D.J.; Gossner, M.M.; Börschig, C.; Blüthgen, N.; de Jong, H.; Simons, N.K.; Klein, A.-M.; Krauss, J.; Maier, G.; et al. Landscape simplification filters species traits and drives biotic homogenization. Nat. Commun. 2015, 6, 8568. [Google Scholar] [CrossRef] [PubMed]
- Violle, C.; Garnier, E.; Lecoeur, J.; Roumet, C.; Podeur, C.; Blanchard, A.; Navas, M.L. Competition, traits and resource depletion in plant communities. Oecologia 2009, 160, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Caruso, C.M.; Remington, D.L.D.; Ostergren, K.E. Variation in resource limitation of plant reproduction influences natural selection on floral traits of Asclepias syriaca. Oecologia 2005, 146, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Kühn, I.; Bierman, S.M.; Durka, W.; Klotz, S. Relating geographical variation in pollination types to environmental and spatial factors using novel statistical methods. New Phytol. 2006, 172, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Van Bodegom, P.M.; Douma, J.C.; Verheijen, L.M. A fully traits-based approach to modeling global vegetation distribution. Proc. Natl. Acad. Sci. USA 2014, 111, 13733–13738. [Google Scholar] [CrossRef] [PubMed]
- Van Bodegom, P.M.; Douma, J.C.; Witte, J.P.M.; Ordoñez, J.C.; Bartholomeus, R.P.; Aerts, R. Going beyond limitations of plant functional types when predicting global ecosystem-atmosphere fluxes: Exploring the merits of traits-based approaches. Glob. Ecol. Biogeogr. 2012, 21, 625–636. [Google Scholar] [CrossRef]
- Diaz, S.; Hodgson, J.G.; Thompson, K.; Cabido, M.; Cornelissen, J.H.C.; Jalili, A.; Montserrat-Marti, G.; Grime, J.P.; Zarrinkamar, F.; Asri, Y.; et al. The plant traits that drive ecosystems: Evidence from three continents. J. Veg. Sci. 2004, 15, 295–304. [Google Scholar] [CrossRef]
- Díaz, S.; Kattge, J.; Cornelissen, J.H.C.; Wright, I.J.; Lavorel, S.; Dray, S.; Reu, B.; Kleyer, M.; Wirth, C.; Prentice, I.C.; et al. The global spectrum of plant form and function. Nature 2015, 529, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.D.; Morsdorf, F.; Schmid, B.; Petchey, O.L.; Hueni, A.; Schimel, D.S.; Schaepman, M.E. Mapping functional diversity from remotely sensed morphological and physiological forest traits. Nat. Commun. 2017, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt-Römermann, M.; Gray, A.; Vanbergen, A.J.; Bergès, L.; Bohner, A.; Brooker, R.W.; De Bruyn, L.; De Cinti, B.; Dirnböck, T.; Grandin, U.; et al. Functional traits and local environment predict vegetation responses to disturbance: A pan-European multi-site experiment. J. Ecol. 2011, 99, 777–787. [Google Scholar] [CrossRef]
- Lavorel, S.; Garnier, E. Predicting changes in community composition and ecosystem functioning from plant traits: Revisting the Holy Grail. Funct. Ecol. 2002, 16, 545–556. [Google Scholar] [CrossRef]
- Reichstein, M.; Bahn, M.; Mahecha, M.D.; Kattge, J.; Baldocchi, D.D. Linking plant and ecosystem functional biogeography. Proc. Natl. Acad. Sci. USA 2014, 111, 13697–13702. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, E.; Altermatt, F.; Cavender-bares, J.; Schuman, M.C.; Zuppinger-dingley, D.; Garonna, I.; Schneider, F.D.; Guille, C.; Van Moorsel, S.J.; Hahl, T.; et al. ScienceDirect Genomics meets remote sensing in global change studies: Monitoring and predicting phenology, evolution and biodiversity. Curr. Opin. Environ. Sustain. 2017, 29, 177–186. [Google Scholar] [CrossRef]
- Spreen, G.; Kaleschke, L.; Heygster, G. Sea ice remote sensing using AMSR-E 89-GHz channels. J. Geophys. Res. Ocean. 2008, 113, 1–14. [Google Scholar] [CrossRef]
- Braun, D.; Damm, A.; Paul-Limoges, E.; Revill, A.; Buchmann, N.; Petchey, O.L.; Hein, L.; Schaepman, M.E. From instantaneous to continuous: Using imaging spectroscopy and in situ data to map two productivity-related ecosystem services. Ecol. Indic. 2017, 82, 409–419. [Google Scholar] [CrossRef]
- Braun, D.; Damm, A.; Petchey, O.L.; Hein, L.; Schaepman, M.E. Spatio-temporal trends and trade-offs in ecosystem services: An Earth observation based assessment for Switzerland between 2004 and 2014. Ecol. Indic. 2017, 89, 828–839. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with Random forest using very high spatial resolution 8-band worldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Lausch, A.; Heurich, M.; Gordalla, D.; Dobner, H.-J.J.; Gwillym-Margianto, S.; Salbach, C. Forecasting potential bark beetle outbreaks based on spruce forest vitality using hyperspectral remote-sensing techniques at different scales. For. Ecol. Manag. 2013, 308, 76–89. [Google Scholar] [CrossRef]
- Deans, A.R.; Lewis, S.E.; Huala, E.; Anzaldo, S.S.; Ashburner, M.; Balhoff, J.P.; Blackburn, D.C.; Blake, J.A.; Burleigh, J.G.; Chanet, B.; et al. Finding Our Way through Phenotypes. PLoS Biol. 2015, 13, e1002033. [Google Scholar] [CrossRef] [PubMed]
- Burleigh, G.; Alphonse, K.; Alverson, A.J.; Bik, H.M.; Blank, C.; Cirranello, A.L.; Cui, H.; Daly, M.; Dietterich, T.G.; Gasparich, G.; et al. Next-generation phenomics for the Tree of Life. PLoS Curr. 2013. [Google Scholar] [CrossRef] [PubMed]
- Deans, A.R.; Yoder, M.J.; Balhoff, J.P. Time to change how we describe biodiversity. Trends Ecol. Evol. 2012, 27, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Bräutigam, K.; Vining, K.J.; Lafon-Placette, C.; Fossdal, C.G.; Mirouze, M.; Marcos, J.G.; Fluch, S.; Fraga, M.F.; Guevara, M.Á.; Abarca, D.; et al. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol. Evol. 2013, 3, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Messier, J.; McGill, B.J.; Lechowicz, M.J. How do traits vary across ecological scales? A case for trait-based ecology. Ecol. Lett. 2010, 13, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, C.M.; Strader, L.C.; Pratt, J.G.; Maiese, D.; Hendershot, T.; Kwok, R.K.; Hammond, J.A.; Huggins, W.; Jackman, D.; Pan, H.; et al. The PhenX toolkit: Get the most from your measures. Am. J. Epidemiol. 2011, 174, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, J.H.C.; Lavorel, S.; Garnier, E.; Díaz, S.; Buchmann, N.; Gurvich, D.E.; Reich, P.B.; Eter, S.; Morgan, H.D.; van der Heijden, M.G.A.; et al. Handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 2003, 51, 335–380. [Google Scholar] [CrossRef]
- Pérez-Harguindeguy, N.; Díaz, S.; Lavorel, S.; Poorter, H.; Jaureguiberry, P.; Bret-Harte, M.S.; Cornwell, W.K.; Craine, J.M.; Gurvich, D.E.; Urcelay, C.; et al. New Handbook for standardized measurment of plant functional traits worldwide. Aust. J. Bot. 2013, 23, 167–234. [Google Scholar] [CrossRef]
- Kattge, J.; Díaz, S.; Lavorel, S.; Prentice, I.C.; Leadley, P.; Bönisch, G.; Garnier, E.; Westoby, M.; Reich, P.B.; Wright, I.J.; et al. TRY—A global database of plant traits. Glob. Chang. Biol. 2011, 17, 2905–2935. [Google Scholar] [CrossRef]
- Schaepman, M.E.; Ustin, S.L.; Plaza, A.J.; Painter, T.H.; Verrelst, J.; Liang, S. Earth system science related imaging spectroscopy—An assessment. Remote Sens. Environ. 2009, 113, S123–S137. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef]
- Ruttenberg, A.; Rees, J.A.; Samwald, M.; Marshall, M.S. Life sciences on the Semantic Web: The Neurocommons and beyond. Brief Bioinform. 2009, 10, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Laporte, M.A.; Mougenot, I.; Garnier, E. ThesauForm-Traits: A web based collaborative tool to develop a thesaurus for plant functional diversity research. Ecol. Inform. 2012, 11, 34–44. [Google Scholar] [CrossRef]
- Garnier, E.; Stahl, U.; Laporte, M.-A.; Kattge, J.; Mougenot, I.; Kühn, I.; Laporte, B.; Amiaud, B.; Ahrestani, F.S.; Bönisch, G.; et al. Towards a thesaurus of plant characteristics: An ecological contribution. J. Ecol. 2017, 105, 298–309. [Google Scholar] [CrossRef]
- Abiven, S.; Altermatt, F.; Backhaus, N.; Deplazes-zemp, A.; Furrer, R.; Korf, B.; Niklaus, P.A.; Schaepman-strub, G.; Shimizu, K.K.; Zuppinger-dingley, D.; et al. ScienceDirect Integrative research efforts at the boundary of biodiversity and global change research. Curr. Opin. Environ. Sustain. 2017, 29, 215–222. [Google Scholar] [CrossRef]
- Ayris, P.; Berthou, J.-Y.; Bruce, R.; Lindstaedt, S.; Monreale, A.; Mons, B.; Murayama, Y.; Södergård, C.; Tochtermann, K.; Wilkinson, R. Realising the European Open Science Cloud; European Union: Brussels, Belgium, 2016. [Google Scholar]
- Abbott, A.; Butler, D.; Gibney, E.; Schiermeier, Q.; Van Noorden, R. Boon or burden: What has the EU ever done for science? Nature 2016, 534, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Freedman, L.P.; Cockburn, I.M.; Simcoe, T.S. The economics of reproducibility in preclinical research. PLoS Biol. 2015, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Read, K.B.; Sheehan, J.R.; Huerta, M.F.; Knecht, L.S.; Mork, J.G.; Humphreys, B.L.; Abhyankar, S.; Akiwumi, O.; Bodenreider, O.; Davidson, S.; et al. Sizing the problem of improving discovery and access to NIH-Funded data: A preliminary study. PLoS ONE 2015, 10, e0132735. [Google Scholar] [CrossRef] [PubMed]
- Wallace, L.; Lucieer, A.; Watson, C.; Turner, D. Development of a UAV-LiDAR system with application to forest inventory. Remote Sens. 2012, 4, 1519–1543. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Naesset, E.; Ørka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef]
- Stysley, P.R.; Coyle, D.B.; Kay, R.B.; Frederickson, R.; Poulios, D.; Cory, K.; Clarke, G. Long term performance of the High Output Maximum Efficiency Resonator (HOMER) laser for NASA’s Global Ecosystem Dynamics Investigation (GEDI) lidar. Opt. Laser Technol. 2015, 68, 67–72. [Google Scholar] [CrossRef]
- Collin, A.; Hench, J.L. Towards Deeper Measurements of Tropical Reefscape Structure Using the WorldView-2 Spaceborne Sensor. Remote Sens. 2012, 4, 1425–1447. [Google Scholar] [CrossRef]
- Woodcock, C.E.; Allen, R.; Anderson, M.; Belward, A.; Bindschadler, R.; Cohen, W.; Gao, F.; Goward, S.N.; Helder, D.; Helmer, E.; et al. Free Access to Landsat Imagery. Science 2008, 320, 1011–1012. [Google Scholar] [CrossRef] [PubMed]
- Cohen, W.B.; Goward, S.N. Landsat’s Role in Ecological Applications of Remote Sensing. Bioscience 2004, 54, 535–545. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Goward, S.N.; Masek, J.G.; Irons, J.R.; Herold, M.; Cohen, W.B.; Loveland, T.R.; Woodcock, C.E. Landsat continuity: Issues and opportunities for land cover monitoring. Remote Sens. Environ. 2008, 112, 955–969. [Google Scholar] [CrossRef]
- Coops, N.C.; Gillanders, S.N.; Wulder, M.A.; Gergel, S.E.; Nelson, T.; Goodwin, N.R. Assessing changes in forest fragmentation following infestation using time series Landsat imagery. For. Ecol. Manag. 2010, 259, 2355–2365. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Guanter, L.; Kaufmann, H.; Segl, K.; Foerster, S.; Rogass, C.; Chabrillat, S.; Kuester, T.; Hollstein, A.; Rossner, G.; Chlebek, C.; et al. The EnMAP spaceborne imaging spectroscopy mission for earth observation. Remote Sens. 2015, 7, 8830–8857. [Google Scholar] [CrossRef]
- Lausch, A.; Schmidt, A.; Tischendorf, L. Data mining and linked open data—New perspectives for data analysis in environmental research. Ecol. Model. 2015, 295, 5–17. [Google Scholar] [CrossRef]
- Bizer, C. The emerging web of linked data. IEEE Intell. Syst. 2009, 24, 87–92. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalversberg, I.J.; Appleton, G.; Axton, M. Comment: The FAIR Guiding Principles for scientific data management and stewardship. Nat. Commun. 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.G.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.M.; Cardoso, A.C.; et al. Essential Biodiversity Variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, E.; Gunn, J.; Fischer, A.; McCurdy, A.; Glover, L.K.; Members, T.T. A Framework for Ocean Observing; UNESCO: Paris, France, 2012. [Google Scholar]
- Bojinski, S.; Verstraete, M.; Peterson, T.C.; Richter, C.; Simmons, A.; Zemp, M. The concept of essential climate variables in support of climate research, applications, and policy. Bull. Am. Meteorol. Soc. 2014, 95, 1431–1443. [Google Scholar] [CrossRef]
- Goerre, S.; Egli, C.; Gerber, S.; Defila, C.; Minder, C.; Richner, H.; Meier, B. Impact of weather and climate on the incidence of acute coronary syndromes. Int. J. Cardiol. 2007, 118, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Scholes, R.J.; Walters, M.; Turak, E.; Saarenmaa, H.; Heip, C.H.; Tuama, É.Ó.; Faith, D.P.; Mooney, H.A.; Ferrier, S.; Jongman, R.H.; et al. Building a global observing system for biodiversity. Curr. Opin. Environ. Sustain. 2012, 4, 139–146. [Google Scholar] [CrossRef]
- GOFC-GOLD. A Sourcebook of Methods and Procedures for Monitoring Essential Biodiversity Variables in Tropical Forests with Remote Sensing; GOFC-GOLD, Bon, Ed.; Wageningen University: Wageningen, The Netherlands, 2017. [Google Scholar]
- Esch, T.; Uereyen, S.; Asamer, H.; Hirner, A.; Marconcini, M.; Metz, A.; Zeidler, J.; Boettcher, M.; Permana, H.; Brito, F.; et al. Earth observation-supported service platform for the development and provision of thematic information on the built environment—The TEP-Urban project. In Proceedings of the 2017 Joint Urban Remote Sensing Event (JURSE), Dubai, United Arab Emirates, 6–8 March 2017. [Google Scholar]
- Ristoski, P.; Bizer, C.; Paulheim, H. Mining the Web of Linked Data with RapidMiner. J. Web Semant. 2015, 35, 142–151. [Google Scholar] [CrossRef]
- Balazinska, M.; Franklin, M.J.; Gibbons, P.B.; Gray, J.; Nath, S.; Hansen, M.; Tao, V. Data Management in the Worldwide Sensor Web. IEEE Pervasive Comput. 2007, 6, 30–40. [Google Scholar] [CrossRef]
- Mazzocchi, F. Could Big Data be the end of theory in science? A few remarks on the epistemology of data-driven science. EMBO Rep. 2015, 16, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
- Boulton, G. The challenges of a Big Data Earth. Big Earth Data 2018, 4471, 1–7. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, H.; Wang, L.; Huang, B.; Ranjan, R.; Zomaya, A.; Jie, W. Remote sensing big data computing: Challenges and opportunities. Future Gener. Comput. Syst. 2015, 51, 47–60. [Google Scholar] [CrossRef]
- Berners-Lee, T.; Hendler, J.; Lassila, O. The Semantic Web. Sci. Am. 2001, 284, 34–43. [Google Scholar] [CrossRef]
- Berners-Lee, T. COMPUTER SCIENCE: Enhanced: Creating a Science of the Web. Science 2006, 313, 769–771. [Google Scholar] [CrossRef] [PubMed]
- Madin, J.S.; Bowers, S.; Schildhauer, M.P.; Jones, M.B. Advancing ecological research with ontologies. Trends Ecol. Evol. 2008, 23, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Morsdorf, F.; Kükenbrink, D.; Schneider, F.D.; Abegg, M.; Schaepman, M.E. Close-range laser scanning in forests: Towards physically based semantics across scales. Interface Focus 2018, 8, 20170046. [Google Scholar] [CrossRef] [PubMed]
- Ristoski, P.; Paulheim, H. Semantic Web in data mining and knowledge discovery: A comprehensive survey. J. Web Semant. 2016, 36, 1–22. [Google Scholar] [CrossRef]
- Pey, B.; Laporte, M.A.; Nahmani, J.; Auclerc, A.; Capowiez, Y.; Caro, G.; Cluzeau, D.; Cortet, J.; Decaëns, T.; Dubs, F.; et al. A thesaurus for soil invertebrate trait-based approaches. PLoS ONE 2014, 9, e108985. [Google Scholar] [CrossRef] [PubMed]
- Paulheim, H. Automatic Knowledge Graph Refinement: A Survey of Approaches and Evaluation Methods. Semant. Web 2015, 8, 489–508. [Google Scholar] [CrossRef]
- Rennolls, K. A partial ontology for forest inventory and mensuration. In Proceedings of the 16th International Workshop on Database and Expert Systems Applications (DEXA’05), Copenhagen, Denmark, 22–26 August 2005; pp. 679–683. [Google Scholar]
- Smith, C.L.; Goldsmith, C.W.; Eppig, J.T. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005, 6, R7. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, P. A Resource for a Common Reference Ontology for Plants. In Proceedings of the Plant and Animal Genome XXI Conference, San Diego, CA, USA, 12 January 2013; pp. 1–2. [Google Scholar]
- Oellrich, A.; Walls, R.L.; Cannon, E.K.; Cannon, S.B.; Cooper, L.; Gardiner, J.; Gkoutos, G.V.; Harper, L.; He, M.; Hoehndorf, R.; et al. An ontology approach to comparative phenomics in plants. Plant Methods 2015, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M. Ontology-based Discovery and Composition of Geographic Information Services. Ph.D. Dissertation, Westfaelischen Wilhelms-Universit, Muenster, Germany, 2005. [Google Scholar]
- Arnaud, E.; Matteis, L.; Laporte, M.A.; Espinosa, H.; Hyman, G.; Shrestha, R.; Portugal, A.; Chibon, P.Y.; Devare, M.; Akintunde, A.; et al. The Crop Ontology, a resource for enabling access to breeders’ data. In Proceedings of the Plant and Animal Genome XXII Conference, San Diego, CA, USA, 14 Januray 2014. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontologie: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Karam, N.; Fichtmueller, D.; Gleisberg, M.; Bohlen, V.; Tolksdorf, R.; Güntsch, A. The Terminology Server of the German Federation for Biological Data (GFBio)—Server and Service of Semantic Technologies in Scientific Environments; GFBio: Berlin, Germany, 2014. [Google Scholar]
- Bush, A.; Sollmann, R.; Wilting, A.; Bohmann, K.; Cole, B.; Balzter, H.; Martius, C.; Zlinszky, A.; Calvignac-Spencer, S.; Cobbold, C.A.; et al. Connecting Earth observation to high-throughput biodiversity data. Nat. Ecol. Evol. 2017, 1, 0176. [Google Scholar] [CrossRef] [PubMed]
- Asadi, A. Advanced Data Mining and Machine Learning Algorithms for Integrated Computer-Based Analyses of Big Environmental Databases; Eberhard Karls University: Tübingen, Germany, 2017. [Google Scholar]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Nemani, R.; Votava, P.; Michaelis, A.; Melton, F.; Milesi, C. Collaborative Supercomputing for Global Change Science. EOS Trans. Am. Geophys. Union 2011, 92, 109. [Google Scholar] [CrossRef]
- Schreiber, L.R.; Bluhm, H. Toward a silicon-based quantum computer. Science 2018, 359, 393–394. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Trivedi, R.K. Literature review: Cloud Computing –Security Issues, Solution and Technologies. Int. J. Eng. Res. 2014, 3, 221–225. [Google Scholar] [CrossRef]
- Xiong, J.; Thenkabail, P.S.; Tilton, J.C.; Gumma, M.K.; Teluguntla, P.; Oliphant, A.; Congalton, R.G.; Yadav, K.; Gorelick, N. Nominal 30-m cropland extent map of continental Africa by integrating pixel-based and object-based algorithms using Sentinel-2 and Landsat-8 data on google earth engine. Remote Sens. 2017, 9, 1065. [Google Scholar] [CrossRef]
- Agapiou, A. Remote sensing heritage in a petabyte-scale: Satellite data and heritage Earth Engine© applications. Int. J. Digit. Earth 2017, 10, 85–102. [Google Scholar] [CrossRef]
- Johansen, K.; Phinn, S.; Taylor, M. Mapping woody vegetation clearing in Queensland, Australia from Landsat imagery using the Google Earth Engine. Remote Sens. Appl. Soc. Environ. 2015, 1, 36–49. [Google Scholar] [CrossRef]
- Aundeen, J.L.F.; Anengieter, R.L.K.; Uswell, M.D.B.U.S. Geological Survey Spatial Data Access. J. Geospat. Eng. 2002, 4, 145–152. [Google Scholar]
- Acker, J.G.; Leptoukh, G. Online Analysis Enhances Use of NASA Earth Science Data. EOS Trans. Am. Geophys. Union 2007, 88, 14–17. [Google Scholar] [CrossRef]
- Wagemann, J.; Clements, O.; Marco Figuera, R.; Rossi, A.P.; Mantovani, S. Geospatial web services pave new ways for server-based on-demand access and processing of Big Earth Data. Int. J. Digit. Earth 2018, 11, 7–25. [Google Scholar] [CrossRef]
- Neitzel, F.; Klonowski, J. Mobile 3D Mapping with a Low-Cost Uav System. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 39–44. [Google Scholar] [CrossRef]
- Eberle, J.; Clausnitzer, S.; Hüttich, C.; Schmullius, C. Multi-Source Data Processing Middleware for Land Monitoring within a Web-Based Spatial Data Infrastructure for Siberia. ISPRS Int. J. Geo-Inform. 2013, 2, 553–576. [Google Scholar] [CrossRef]
- Hardisty, A.R.; Bacall, F.; Beard, N.; Balcázar-Vargas, M.-P.; Balech, B.; Barcza, Z.; Bourlat, S.J.; De Giovanni, R.; de Jong, Y.; De Leo, F.; et al. BioVeL: A virtual laboratory for data analysis and modelling in biodiversity science and ecology. BMC Ecol. 2016, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- McIlraith, S.A.; Son, T.C.; Zeng, H. Semantic Web services. IEEE Intell. Syst. 2001, 16, 46–53. [Google Scholar] [CrossRef]
- Martin, D.; Paolucci, M.; McIlraith, S.; Burstein, M.; McDermott, D.; McGuinness, D.; Parsia, B.; Payne, T.; Sabou, M.; Solanki, M.; et al. Bringing Semantics to Web Services: The OWL-S Approach; Springer: Berlin/Heidelberg, Germany, 2005; pp. 26–42. [Google Scholar]
- Sudmanns, M.; Tiede, D.; Lang, S.; Baraldi, A. Semantic and syntactic interoperability in online processing of big Earth observation data. Int. J. Digit. Earth 2018, 11, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Kiker, G.A.; Bridges, T.S.; Varghese, A.; Seager, T.P.; Linkov, I. Application of Multicriteria Decision Analysis in Environmental Decision Making. Integr. Environ. Assess. Manag. 2005, 1, 95. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Demissew, S.; Carabias, J.; Joly, C.; Lonsdale, M.; Ash, N.; Larigauderie, A.; Adhikari, J.R.; Arico, S.; Báldi, A.; et al. The IPBES Conceptual Framework—Connecting nature and people. Curr. Opin. Environ. Sustain. 2015, 14, 1–16. [Google Scholar] [CrossRef]
- Jelokhani-Niaraki, M.; Sadeghi-Niaraki, A.; Choi, S.M. Semantic interoperability of GIS and MCDA tools for environmental assessment and decision making. Environ. Model. Softw. 2018, 100, 104–122. [Google Scholar] [CrossRef]
- Altintas, I.; Berkley, C.; Jaeger, E.; Jones, M.; Ludascher, B.; Mock, S. Kepler: An extensible system for design and execution of scientific workflows. In Proceedings of the 16th International Conference on Scientific and Statistical Database Management, Santorini Island, Greece, 21–23 June 2004; pp. 423–424. [Google Scholar]
- Deelman, E.; Vahi, K.; Juve, G.; Rynge, M.; Callaghan, S.; Maechling, P.J.; Mayani, R.; Chen, W.; Ferreira da Silva, R.; Livny, M.; et al. Pegasus, a workflow management system for science automation. Future Gener. Comput. Syst. 2015, 46, 17–35. [Google Scholar] [CrossRef]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Wiswedel, B. KNIME—The Konstanz information miner: Version 2.0 and beyond. AcM SIGKDD Explor. Newsl. 2009, 11, 26–31. [Google Scholar] [CrossRef]
- Hofmann, M.; Klinkenberg, R. (Eds.) RapidMiner: Data Mining Use Cases and Business Analytics Applications; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Oinn, T.; Addis, M.; Ferris, J.; Marvin, D.; Senger, M.; Greenwood, M.; Carver, T.; Glover, K.; Pocock, M.R.; Wipat, A.; et al. Taverna: A tool for the composition and enactment of bioinformatics workflows. Bioinformatics 2004, 20, 3045–3054. [Google Scholar] [CrossRef] [PubMed]
- Wolstencroft, K.; Haines, R.; Fellows, D.; Williams, A.; Withers, D.; Owen, S.; Soiland-Reyes, S.; Dunlop, I.; Nenadic, A.; Fisher, P.; et al. The Taverna workflow suite: Designing and executing workflows of Web Services on the desktop, web or in the cloud. Nucleic Acids Res. 2013, 41, 557–561. [Google Scholar]
- Callahan, S.P.; Freire, J.; Santos, E.; Scheidegger, C.E.; Silva, C.T.; Vo, H.T. Managing the Evolution of Dataflows with VisTrails. In Proceedings of the 22nd International Conference on Data Engineering Workshops (ICDEW’06), Atlanta, GA, USA, 3–7 April 2006; p. 71. [Google Scholar]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; Afgan, E.; Ananda, G.; Baker, D.; Blankenberg, D.; Chakrabarty, R.; Coraor, N.; Goecks, J.; et al. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef] [PubMed]
- Bizer, C.; Heath, T.; Berners-Lee, T. Linked data-the story so far. Int. J. Semant. Web Inf. Syst. 2009, 5, 1–22. [Google Scholar]
- Zhao, Z.; Martin, P.; Grosso, P.; Los, W.; De Laat, C.; Jeffrey, K.; Hardisty, A.; Vermeulen, A.; Castelli, D.; Legre, Y.; et al. Reference model guided system design and implementation for interoperable environmental research infrastructures. In Proceedings of the 11th IEEE International Conference eScience, Munich, Germany, 31 August–4 September 2015; pp. 551–556. [Google Scholar]













| Level/Scale | Responsible Body | Description | Resources/Links |
|---|---|---|---|
| Country level: Germany | Johann Heinrich von Thünen-Institute, Federal Research Institute for Rural Areas, Forestry and Fisheries in cooperation with other institutions (Thünen Institute of Forest Ecosystems, Thünen Institute of International Forestry and Forest Economics) | Forest condition monitoring (FCM) Level-I Monitoring Frequency: annual Compilation of national reports on forest conditions for Germany (FCA—Forest Conservation Act). Forest health (FH) assessed using systematic sample grid of permanent plots. | [35] |
| Johann Heinrich von Thünen-Institute, Federal Research Institute for Rural Areas, Forestry and Fisheriesin cooperation with other institutions (Thünen Institute of Forest Ecosystems, Thünen Institute of International Forestry and Forest Economics) | Intensive monitoring Level-II Monitoring Frequency: continuous 66 sites intensively monitored, partly through continual sampling of relevant ecosystem compartments in selected forest ecosystems. | [36] | |
| Johann Heinrich von Thünen-Institute, Federal Research Institute for Rural Areas, Forestry and Fisheries in cooperation with the Federal Ministry for Food and Agriculture supported by the European Union (EU) and the Food and Agriculture Organization of the United Nations (FAO) | National Forest Inventory (NFI) Level-III Monitoring Frequency: every 10 years Status and development of the forests of Germany derived from a sample-based large-scale forest inventory 45 countries are integrated into the NFI. | [37] | |
| Country level: USA | United States Department of Agriculture(USDA) Forest Service | FH Monitoring (M) Frequency: annual National program designed to determine the status, changes, and trends in indicators of forest condition on an annual basis. | [38] |
| Country level: China | State Forestry Administration of China | National Forest Inventory including the protocol for Monitoring Forest Health Frequency: five years Since 2004, components that monitored the health status and ecological functions of forest ecosystems were added to the NFI. | [6,39] |
| Country level: Canada | Canadian Forest Service (CFS) | National FHM Network Frequency: 5-years Established in 1994 based on earlier Acid Rain Monitoring Network. Plot-based biannual to five-year repetition depending on the variables. | [40] |
| National Forest Inventory (NFI), Canada | National Forest Inventory Frequency: 10 years Mixed plot and remote sensing (RS)-based. Includes inventory parameters and assessment of insect, disease, fire, and other disturbance damage. | [41] | |
| European level | United Nations Economic Commission for Europe (UNECE) | ICP Forest: the International Co-operative Programme on Assessment and Monitoring of Air Pollution Effects on Forests Frequency: annual Level 1: Long-term monitoring, 6000 sampling sites in Europe with 16 km2 Level 2: Long-term intensive sites, for process-orientated study Effects on Forest. Developed to standardize the recording of different FH indicators on three levels of intensity. ICP Forest Manuals: Development of standardized measurement protocols http://icp-forests.net/page/icp-forests-manual | [42] |
| European Union (EU) | FOREST EUROPE-Program | European Union (EU) | |
| Global level (45 countries) | Food and Agriculture Organization of the United Nations (FAO) | National Forest Inventory (NFI) Frequency: every 10 years; 45 countries are integrated into the NFI | [41] |
| Global level | Food and Agriculture Organization of the United Nations (FAO) | Forest Resources Assessment Frequency: five yearsFH recorded by the FAO as part of the Forest Resources Assessment (FRA). Individual countries report their findings to the FAO, which then compiles a report. FAO FRA Terms and Definitions: Development of standardized measurement protocols (http://www.fao.org/docrep/017/ap862e/ap862e00.pdf) | [43] |
| Harmonization Initiatives | |||
| European level | Forest Information System for Europe—(FISE) | http://data.jrc.ec.europa.eu/collection/FISE; http://fise.jrc.ec.europa.eu/) | |
| Global level | Global Forest Observations Initiative (GFOI) | (http://www.gfoi.org/about-gfoi/) | |
| The Food and Agriculture Organization (FAO) | Global Forest SurveyEstablish a global network of systematic sampling plots to understand forest ecosystems’ characteristics and health status, and create a global web-based platform where forest data are available and freely shared. | http://www.fao.org/in-action/global-forest-survey/en/ | |
| Acronym | Name | Link | Reference |
|---|---|---|---|
| USFS FIA program | United States Forest Service (USFS) Forest Inventory and Analysis (FIA) database | https://www.fia.fs.fed.us/ https://www.fia.fs.fed.us/tools-data/index.php | [59,60] |
| TreeDivNET | Global network of tree diversity experiments | http://www.treedivnet.ugent.be/ | [61] |
| T4F | Trees4Future | http://www.trees4future.eu/database-access.html | [62] |
| FunDivEUROPE Exploratory Platform | Functional significance of forest biodiversity | http://project.fundiveurope.eu/ | [63] |
| BIOTREE-NET | BIOTREE—BIOdiversity and ecosystem processes in experimental TREE stands | http://portal.biotreenet.com/ | [10,64] |
| Global Forest Watch | Global Forest Watch | http://www.globalforestwatch.org/ | [65,66] |
| GFOI | Global Forest Observations Initiative | http://www.gfoi.org/ | [66] |
| GENFORED | Forest genetic trials network | https://sites.cita-aragon.es/genfored/en/welcome-to-the-genfored-web-site/ | |
| EUFORGEN | European forest genetic resources program | http://www.euforgen.org/ | [67] |
| Characteristics | Phenotypes | Traits |
|---|---|---|
| Indicators and filters of status, stress, disturbances, and resource limitations. | [160] | [24] |
| Can be recorded on all levels of biological organization, as well as in different domains. | [160,161] | [164] |
| Standardized measurements are possible. | [165] | [166,167,168,169,170] |
| Can be transferred through a human–computer communication interpretable language in digital form, and can be integrated into the entire system. | [160,171] | [172,173] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lausch, A.; Borg, E.; Bumberger, J.; Dietrich, P.; Heurich, M.; Huth, A.; Jung, A.; Klenke, R.; Knapp, S.; Mollenhauer, H.; et al. Understanding Forest Health with Remote Sensing, Part III: Requirements for a Scalable Multi-Source Forest Health Monitoring Network Based on Data Science Approaches. Remote Sens. 2018, 10, 1120. https://doi.org/10.3390/rs10071120
Lausch A, Borg E, Bumberger J, Dietrich P, Heurich M, Huth A, Jung A, Klenke R, Knapp S, Mollenhauer H, et al. Understanding Forest Health with Remote Sensing, Part III: Requirements for a Scalable Multi-Source Forest Health Monitoring Network Based on Data Science Approaches. Remote Sensing. 2018; 10(7):1120. https://doi.org/10.3390/rs10071120
Chicago/Turabian StyleLausch, Angela, Erik Borg, Jan Bumberger, Peter Dietrich, Marco Heurich, Andreas Huth, András Jung, Reinhard Klenke, Sonja Knapp, Hannes Mollenhauer, and et al. 2018. "Understanding Forest Health with Remote Sensing, Part III: Requirements for a Scalable Multi-Source Forest Health Monitoring Network Based on Data Science Approaches" Remote Sensing 10, no. 7: 1120. https://doi.org/10.3390/rs10071120