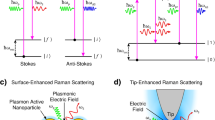
Abstract
Biofilms represent the predominant form of microbial life on our planet. These aggregates of microorganisms, which are embedded in a matrix formed by extracellular polymeric substances, may colonize nearly all interfaces. Detailed knowledge of microorganisms enclosed in biofilms as well as of the chemical composition, structure, and functions of the complex biofilm matrix and their changes at different stages of the biofilm formation and under various physical and chemical conditions is relevant in different fields. Important research topics include the development and improvement of antibiotics and medical devices and the optimization of biocides, antifouling strategies, and biological wastewater treatment. Raman microspectroscopy is a capable and nondestructive tool that can provide detailed two-dimensional and three-dimensional chemical information about biofilm constituents with the spatial resolution of an optical microscope and without interference from water. However, the sensitivity of Raman microspectroscopy is rather limited, which hampers the applicability of Raman microspectroscopy especially at low biomass concentrations. Fortunately, the resonance Raman effect as well as surface-enhanced Raman scattering can help to overcome this drawback. Furthermore, the combination of Raman microspectroscopy with other microscopic techniques, mass spectrometry techniques, or particularly with stable-isotope techniques can provide comprehensive information on monospecies and multispecies biofilms. Here, an overview of different Raman microspectroscopic techniques, including resonance Raman microspectroscopy and surface-enhanced Raman scattering microspectroscopy, for in situ detection, visualization, identification, and chemical characterization of biofilms is given, and the main feasibilities and limitations of these techniques in biofilm research are presented. Future possibilities of and challenges for Raman microspectroscopy alone and in combination with other analytical techniques for characterization of complex biofilm matrices are discussed in a critical review.

Applicability of Raman microspectroscopy for biofilm analysis










Similar content being viewed by others
References
Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–22.
Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108.
Flemming HC. Biofouling in water systems - cases, causes and countermeasures. Appl Microbiol Biotechnol. 2002;59(6):629–40.
An D, Parsek MR. The promise and peril of transcriptional profiling in biofilm communities. Curr Opin Microbiol. 2007;10(3):292–6.
Stewart PS, William CJ. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–8.
Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13(1):20–6.
Sandt C, Smith-Palmer T, Pink J, Brennan L, Pink D. Confocal Raman microspectroscopy as a tool for studying the chemical heterogeneities of biofilms in situ. J Appl Microbiol. 2007;103(5):1808–20.
Ivleva NP, Wagner M, Horn H, Niessner R, Haisch C. Towards a nondestructive chemical characterization of biofilm matrix by Raman microscopy. Anal Bioanal Chem. 2009;393(1):197–206.
Andrews JS, Rolfe SA, Huang WE, Scholes JD, Banwart SA. Biofilm formation in environmental bacteria is influenced by different macromolecules depending on genus and species. Environ Microbiol. 2010;12(9):2496–507.
Chen Y-P, Zhang P, Guo J-S, Fang F, Gao X, Li C. Functional groups characteristics of EPS in biofilm growing on different carriers. Chemosphere. 2013;92(6):633–8.
Lu X, Samuelson DR, Rasco BA, Konkel ME. Antimicrobial effect of diallyl sulphide on Campylobacter jejuni biofilms. J Antimicrob Chemother. 2012;67(8):1915–26.
Jung GB, Nam SW, Choi S, Lee G-J, Park H-K. Evaluation of antibiotic effects on Pseudomonas aeruginosa biofilm using Raman spectroscopy and multivariate analysis. Biomed Opt Express. 2014;5(9):3238–51.
Prabhawathi V, Boobalan T, Sivakumar PM, Doble M. Functionalized polycaprolactam as an active food package for antibiofilm activity and extended shelf life. Colloids Surf B. 2014;123:461–8.
Kniggendorf A-K, Nogueira R, Kelb C, Schadzek P, Meinhardt-Wollweber M, Ngezahayo A, et al. Confocal Raman microscopy and fluorescent in situ hybridization - a complementary approach for biofilm analysis. Chemosphere. 2016;161:112–8.
Schwartz T, Jungfer C, Heißler S, Friedrich F, Faubel W, Obst U. Combined use of molecular biology taxonomy, Raman spectrometry, and ESEM imaging to study natural biofilms grown on filter materials at waterworks. Chemosphere. 2009;77(2):249–57.
Wagner M, Ivleva NP, Haisch C, Niessner R, Horn H. Combined use of confocal laser scanning microscopy (CLSM) and Raman microscopy (RM): investigations on EPS - matrix. Water Res. 2009;43(1):63–76.
Feng J, de la Fuente-Nunez C, Trimble MJ, Xu J, Hancock REW, Lu X. An in situ Raman spectroscopy-based microfluidic "lab-on-a-chip" platform for non-destructive and continuous characterization of Pseudomonas aeruginosa biofilms. Chem Commun. 2015;51(43):8966–9.
Masyuko RN, Lanni EJ, Driscoll CM, Shrout JD, Sweedler JV, Bohn PW. Spatial organization of Pseudomonas aeruginosa biofilms probed by combined matrix-assisted laser desorption ionization mass spectrometry and confocal Raman microscopy. Analyst. 2014;139(22):5700–8.
Lanni EJ, Masyuko RN, Driscoll CM, Dunham SJB, Shrout JD, Bohn PW, et al. Correlated imaging with C(60)-SIMS and confocal Raman microscopy: visualization of cell-scale molecular distributions in bacterial biofilms. Anal Chem. 2014;86(21):10885–91.
Janissen R, Murillo DM, Niza B, Sahoo PK, Nobrega MM, Cesar CL, et al. Spatiotemporal distribution of different extracellular polymeric substances and filamentation mediate Xylella fastidiosa adhesion and biofilm formation. Sci Rep. 2015;5.
Pätzold R, Keuntje M, Anders-von AA. A new approach to non-destructive analysis of biofilms by confocal Raman microscopy. Anal Bioanal Chem. 2006;386(2):286–92.
Kniggendorf A-K, Meinhardt-Wollweber M. Of microparticles and bacteria identification – (resonance) Raman micro-spectroscopy as a tool for biofilm analysis. Water Res. 2011;45(15):4571–82.
Ivleva NP, Wagner M, Horn H, Niessner R, Haisch C. In situ surface-enhanced Raman scattering analysis of biofilm. Anal Chem. 2008;80(22):8538–44.
Ivleva NP, Wagner M, Szkola A, Horn H, Niessner R, Haisch C. Label-free in situ SERS imaging of biofilms. J Phys Chem B. 2010;114(31):10184–94.
Ivleva NP, Wagner M, Horn H, Niessner R, Haisch C. Raman microscopy and surface-enhanced Raman scattering (SERS) for in situ analysis of biofilms. J Biophotonics. 2010;3(8-9):548–56.
Ramya S, George RP, Rao RVS, Dayal RK. Detection of algae and bacterial biofilms formed on titanium surfaces using micro-Raman analysis. Appl Surf Sci. 2010;256(16):5108–15.
Chao Y, Zhang T. Surface-enhanced Raman scattering (SERS) revealing chemical variation during biofilm formation: from initial attachment to mature biofilm. Anal Bioanal Chem. 2012;404(5):1465–75.
Efeoglu E, Culha M. In situ monitoring of biofilm formation by using surface-enhanced Raman scattering. Appl Spectrosc. 2013;67(5):498–505.
Efeoglu E, Culha M. Surface-enhanced Raman scattering for biofilm characterization. Spectroscopy. 2013;28(11):36–41.
Marcotte L, Barbeau J, Lafleur M. Characterization of the diffusion of polyethylene glycol in Streptococcus mutans biofilms by Raman microspectroscopy. Appl Spectrosc. 2004;58(11):1295–301.
Choo-Smith LP, Maquelin K, Van Vreeswijk T, Bruining HA, Puppels GJ, Thi NAN, et al. Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl Environ Microbiol. 2001;67(4):1461–9.
Sandt C, Smith-Palmer T, Comeau J, Pink D. Quantification of water and biomass in small colony variant PAO1 biofilms by confocal Raman microspectroscopy. Appl Microbiol Biotechnol. 2009;83(6):1171–82.
Samek O, Al-Marashi JFM, Telle HH. The potential of Raman spectroscopy for the identification of biofilm formation by Staphylococcus epidermidis. Laser Phys Lett. 2010;7(5):378–83.
Beier B, Quivey R, Berger A. Raman microspectroscopy for species identification and mapping within bacterial biofilms. AMB Express. 2012;2(1):1–6.
Huang WE, Bailey MJ, Thompson IP, Whiteley AS, Spiers AJ. Single-cell Raman spectral profiles of Pseudomonas fluorescens SBW25 reflects in vitro and in planta metabolic history. Microb Ecol. 2007;53(3):414-25.
Huang WE, Ude S, Spiers AJ. Pseudomonas fluorescens SBW25 biofilm and planktonic cells have differentiable Raman spectral profiles. Microb Ecol. 2007;53(3):471–4.
Kusić D, Kampe B, Ramoji A, Neugebauer U, Rösch P, Popp J. Raman spectroscopic differentiation of planktonic bacteria and biofilms. Anal Bioanal Chem. 2015;1–11.
Smith-Palmer T, Lin S, Oguejiofor I, Leng T, Pustam A, Yang J, et al. In situ confocal Raman microscopy of hydrated early stages of bacterial biofilm formation on various surfaces in a flow cell. Appl Spectrosc. 2016;70(2):289–301.
Virdis B, Harnisch F, Batstone DJ, Rabaey K, Donose BC. Non-invasive characterization of electrochemically active microbial biofilms using confocal Raman microscopy. Energy Environ Sci. 2012;5(5):7017–24.
Virdis B, Millo D, Donose BC, Batstone DJ. Real-time measurements of the redox states of c-type cytochromes in electroactive biofilms: a confocal resonance Raman microscopy study. PLoS One. 2014;9(2), e89918.
Lebedev N, Strycharz-Glaven SM, Tender LM. Spatially resolved confocal resonant Raman microscopic analysis of anode-grown Geobacter sulfurreducens biofilms. ChemPhysChem. 2014;15(2):320–7.
Chen P, Cui L, Zhang K. Surface-enhanced Raman spectroscopy monitoring the development of dual-species biofouling on membrane surfaces. J Membr Sci. 2015;473:36–44.
Cui L, Chen P, Zhang B, Zhang D, Li J, Martin FL, et al. Interrogating chemical variation via layer-by-layer SERS during biofouling and cleaning of nanofiltration membranes with further investigations into cleaning efficiency. Water Res. 2015;87:282–91.
Garima S, Alka P. Combined use of Fourier transform infrared and Raman spectroscopy to study planktonic and biofilm cells of Cronobacter sakazakii. J Microbiol Biotechnol Food Sci. 2014;3(4):310–4.
Noothalapati Venkata HN, Nomura N, Shigeto S. Leucine pools in Escherichia coli biofilm discovered by Raman imaging. J Raman Spectrosc. 2011;42(11):1913–5.
Suci PA, Geesey GG, Tyler BJ. Integration of Raman microscopy, differential interference contrast microscopy, and attenuated total reflection Fourier transform infrared spectroscopy to investigate chlorhexidine spatial and temporal distribution in Candida albicans biofilms. J Microbiol Methods. 2001;46(3):193–208.
Kusić D, Rösch P, Popp J. Fast label-free detection of Legionella spp. in biofilms by applying immunomagnetic beads and Raman spectroscopy. Syst Appl Microbiol. 2016;39(2):132–40.
Liu H, Xu Q, Huo L, Wei X, Ling J. Chemical composition of Enterococcus faecalis in biofilm cells initiated from different physiologic states. Folia Microbiol. 2014;59(5):447–53.
Pätzold R, Keuntje M, Theophile K, Mueller J, Mielcarek E, Ngezahayo A, et al. In situ mapping of nitrifiers and anammox bacteria in microbial aggregates by means of confocal resonance Raman microscopy. J Microbiol Methods. 2008;72(3):241–8.
Sandt C, Smith-Palmer T, Pink J, Pink D. A confocal Raman microscopy study of the distribution of a carotene-containing yeast in a living Pseudomonas aeruginosa biofilm. Appl Spectrosc. 2008;62(9):975–83.
Kögler M, Zhang B, Cui L, Shi Y, Yliperttula M, Laaksonen T, et al. Real-time Raman based approach for identification of biofouling. Sensors Actuators B. 2016;230:411–21.
Millo D, Harnisch F, Patil SA, Ly HK, Schröder U, Hildebrandt P. In situ spectroelectrochemical investigation of electrocatalytic microbial biofilms by surface-enhanced resonance Raman spectroscopy. Angew Chem Int Ed. 2011;50(11):2625–7.
Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11(2):94–100.
Vertes A, Hitchins V, Phillips KS. Analytical challenges of microbial biofilms on medical devices. Anal Chem. 2012;84(9):3858–66.
Cam D, Keseroglu K, Kahraman M, Sahin F, Culha M. Multiplex identification of bacteria in bacterial mixtures with surface-enhanced Raman scattering. J Raman Spectrosc. 2010;41(5):484–9.
Thurnheer T, Gmür R, Guggenheim B. Multiplex FISH analysis of a six-species bacterial biofilm. J Microbiol Methods. 2004;56(1):37–47.
Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol. 2007;9(8):1878–89.
Neu TR, Lawrence JR. In situ characterization of extracellular polymeric substances (EPS) in biofilm systems. Microb Extracell Polym Subst. 1999;21–47.
Lawrence JR, Neu TR, Swerhone GDW. Application of multiple parameter imaging for the quantification of algal, bacterial and exopolymer components of microbial biofilms. J Microbiol Methods. 1998;32(3):253–61.
Alhede M, Qvortrup K, Liebrechts R, Hoiby N, Givskov M, Bjarnsholt T. Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition. FEMS Immunol Med Microbiol. 2012;65(2):335–42.
Haisch C, Niessner R. Visualisation of transient processes in biofilms by optical coherence tomography. Water Res. 2007;41(11):2467–72.
Li C, Felz S, Wagner M, Lackner S, Horn H. Investigating biofilm structure developing on carriers from lab-scale moving bed biofilm reactors based on light microscopy and optical coherence tomography. Bioresour Technol. 2016;200:128–36.
Wright CJ, Shah MK, Powell LC, Armstrong I. Application of AFM from microbial cell to biofilm. Scanning. 2010;32(3):134–49.
Schmid T, Panne U, Haisch C, Hausner M, Niessner R. A photoacoustic technique for depth-resolved in situ monitoring of biofilms. Environ Sci Technol. 2002;36(19):4135–41.
Reichhardt C, Fong JCN, Yildiz F, Cegelski L. Characterization of the Vibrio cholerae extracellular matrix: a top-down solid-state NMR approach. Biochim Biophys Acta. 2015;1848(1 Part B):378–83.
Beier BD, Berger AJ. Method for automated background subtraction from Raman spectra containing known contaminants. Analyst. 2009;134(6):1198–202.
Rae A, Stosch R, Klapetek P, Hight Walker AR, Roy D. State of the art Raman techniques for biological applications. Methods. 2014;68(2):338–47.
Palonpon AF, Ando J, Yamakoshi H, Dodo K, Sodeoka M, Kawata S, et al. Raman and SERS microscopy for molecular imaging of live cells. Nat Protoc. 2013;8(4):677–92.
Hong W, Liao C-S, Zhao H, Younis W, Zhang Y, Seleem MN, et al. In situ detection of a single bacterium in complex environment by hyperspectral CARS imaging. ChemistrySelect. 2016;1(3):513–7.
Schuster KC, Urlaub E, Gapes JR. Single-cell analysis of bacteria by Raman microscopy: spectral information on the chemical composition of cells and on the heterogeneity in a culture. J Microbiol Methods. 2000;42(1):29–38.
Rösch P, Harz M, Schmitt M, Peschke K-D, Ronneberger O, Burkhardt H, et al. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl Environ Microbiol. 2005;71(3):1626–37.
Kumar V, Kampe B, Rösch P, Popp J. Classification and identification of pigmented cocci bacteria relevant to the soil environment via Raman spectroscopy. Environ Sci Pollut Res. 2015;1–9.
Münchberg U, Rösch P, Bauer M, Popp J. Raman spectroscopic identification of single bacterial cells under antibiotic influence. Anal Bioanal Chem. 2014;406(13):3041–50.
Harz M, Rösch P, Popp J. Vibrational spectroscopy - a powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry A. 2009;75A(2):104–13.
Maquelin K, Kirschner C, Choo-Smith L-P, Ngo-Thi NA, van Vreeswijk T, Stämmler M, et al. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J Clin Microbiol. 2003;41(1):324–9.
Maquelin K, Choo-Smith L-P, Endtz HP, Bruining HA, Puppels GJ. Rapid identification of Candida species by confocal Raman microspectroscopy. J Clin Microbiol. 2002;40(2):594–600.
Pahlow S, Meisel S, Cialla-May D, Weber K, Rösch P, Popp J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv Drug Deliv Rev. 2015;89:105–20.
Spiro TG, Strekas TC. Resonance Raman spectra of heme proteins. effects of oxidation and spin state. J Am Chem Soc. 1974;96(2):338–45.
Salama S, Spiro TG. Visible and near-ultraviolet resonance Raman spectra of photolabile vitamin B12 derivatives with a rapid-flow technique. J Raman Spectrosc. 1977;6(2):57–60.
Lutz M. Resonance Raman spectra of chlorophyll in solution. J Raman Spectrosc. 1974;2(5):497–516.
Li M, Canniffe DP, Jackson PJ, Davison PA, FitzGerald S, Dickman MJ, et al. Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J. 2012;6(4):875–85.
Palings I, Pardoen JA, Van den Berg E, Winkel C, Lugtenburg J, Mathies RA. Assignment of fingerprint vibrations in the resonance Raman spectra of rhodopsin, isorhodopsin, and bathorhodopsin: implications for chromophore structure and environment. Biochemistry. 1987;26(9):2544–56.
Copeland RA, Spiro TG. Ultraviolet resonance Raman spectroscopy of flavin mononucleotide and flavin-adenine dinucleotide. J Phys Chem. 1986;90(25):6648–54.
Le Ru EC, Etchegoin PG. Single-molecule surface-enhanced Raman spectroscopy. Annu Rev Phys Chem. 2012;63(1):65–87.
Etchegoin PG, Le Ru EC. A perspective on single molecule SERS: current status and future challenges. Phys Chem Chem Phys. 2008;10(40):6079–89.
Guerrini L, Graham D. Molecularly-mediated assemblies of plasmonic nanoparticles for surface-enhanced Raman spectroscopy applications. Chem Soc Rev. 2012;41(21):7085–107.
Zeiri L, Efrima S. Surface-enhanced Raman spectroscopy of bacteria: the effect of excitation wavelength and chemical modification of the colloidal milieu. J Raman Spectrosc. 2005;36(6/7):667–75.
Schlücker S. Surface-enhanced Raman spectroscopy: concepts and chemical applications. Angew Chem Int Ed. 2014;53(19):4756–95.
Etchegoin PG, Le Ru EC. Basic electromagnetic theory of SERS. In: Schlücker S, editor. Surface enhanced Raman spectroscopy. Weinheim: Wiley-VCH; 2010. p. 1–37.
Bell SEJ, Stewart A. Quantitative SERS methods. In: Schlücker S, editors. Surface enhanced Raman spectroscopy. Weinheim: Wiley-VCH; 2010. p. 71–86.
Otto A. On the significance of Shalaev's ‘hot spots’ in ensemble and single-molecule SERS by adsorbates on metallic films at the percolation threshold. J Raman Spectrosc. 2006;37(9):937–47.
Picorel R, Holt RE, Cotton TM, Seibert M. Surface-enhanced resonance Raman scattering spectroscopy of bacterial photosynthetic membranes. The carotenoid of Rhodospirillum rubrum. J Biol Chem. 1988;263(9):4374–80.
Efrima S, Bronk BV. Silver colloids impregnating or coating bacteria. J Phys Chem B. 1998;102(31):5947–50.
Jarvis RM, Goodacre R. Characterisation and identification of bacteria using SERS. Chem Soc Rev. 2008;37(5):931–6.
Efrima S, Zeiri L. Understanding SERS of bacteria. J Raman Spectrosc. 2009;40(3):277–88.
Ranjith Premasiri W, Lemler P, Chen Y, Gebregziabher Y, Ziegler LD. SERS analysis of bacteria, human blood, and cancer cells: a metabolomic and diagnostic Tool. In: Ozaki Y, Kneipp K, Aroca R, editors. Frontiers of surface-enhanced Raman scattering. Chichester: Wiley; 2014. p. 257–83.
Pearman WF, Lawrence-Snyder M, Angel SM, Decho AW. Surface-enhanced Raman spectroscopy for in situ measurements of signaling molecules (autoinducers) relevant to bacteria quorum sensing. Appl Spectrosc. 2007;61(12):1295–300.
Bodelon G, Montes-Garcia V, Lopez-Puente V, Hill EH, Hamon C, Sanz-Ortiz MN, et al. Detection and imaging of quorum sensing in Pseudomonas aeruginosa biofilm communities by surface-enhanced resonance Raman scattering. Nat Mater. 2016;15(11):1203–11.
Kahraman M, Keseroglu K, Culha M. On sample preparation for surface-enhanced Raman scattering (SERS) of bacteria and the source of spectral features of the spectra. Appl Spectrosc. 2011;65(5):500–6.
Zeiri L, Bronk BV, Shabtai Y, Czege J, Efrima S. Silver metal induced surface enhanced Raman of bacteria. Colloids Surf A Physicochem Eng Asp. 2002;208(1-3):357–62.
Kubryk P, Niessner R, Ivleva NP. The origin of the band at around 730 cm-1 in the SERS spectra of bacteria: a stable isotope approach. Analyst. 2016;141(10):2874–8.
Premasiri WR, Lee JC, Sauer-Budge A, Théberge R, Costello CE, Ziegler LD. The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS. Anal Bioanal Chem. 2016;408(17):4631–47.
Jarvis RM, Law N, Shadi IT, O'Brien P, Lloyd JR, Goodacre R. Surface-enhanced Raman scattering from intracellular and extracellular bacterial locations. Anal Chem. 2008;80(17):6741–6.
Kahraman M, Yazici MM, Sahin F, Bayrak OF, Culha M. Reproducible surface-enhanced Raman scattering spectra of bacteria on aggregated silver nanoparticles. Appl Spectrosc. 2007;61(5):479–85.
Knauer M, Ivleva NP, Niessner R, Haisch C. Optimized SERS colloids for the characterization of microorganisms. Anal Sci. 2010;26:761–6.
Kahraman M, Zamaleeva AI, Fakhrullin RF, Culha M. Layer-by-layer coating of bacteria with noble metal nanoparticles for surface-enhanced Raman scattering. Anal Bioanal Chem. 2009;395(8):2559–67.
Kahraman M, Yazici MM, Sahin F, Culha M. Convective assembly of bacteria for surface-enhanced Raman scattering. Langmuir. 2008;24(3):894–901.
Zhou H, Yang D, Ivleva NP, Mircescu NE, Niessner R, Haisch C. SERS detection of bacteria in water by in situ coating with Ag nanoparticles. Anal Chem. 2014;86(3):1525–33.
Liu X, Knauer M, Ivleva NP, Niessner R, Haisch C. Synthesis of core-shell surface-enhanced Raman tags for bioimaging. Anal Chem. 2010;82(1):441–6.
Jarvis RM, Goodacre R. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal Chem. 2004;76(1):40–7.
Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones II G, Ziegler LD. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J Phys Chem B. 2005;109(1):312–20.
Szeghalmi A, Kaminskyj S, Roesch P, Popp J, Gough KM. Time fluctuations and imaging in the SERS spectra of fungal Hypha grown on nanostructured substrates. J Phys Chem B. 2007;111(44):12916–24.
Neugebauer U, Roesch P, Schmitt M, Popp J, Julien C, Rasmussen A, et al. On the way to nanometer-sized information of the bacterial surface by tip-enhanced Raman spectroscopy. ChemPhysChem. 2006;7(7):1428–30.
Neugebauer U, Schmid U, Baumann K, Ziebuhr W, Kozitskaya S, Deckert V, et al. Towards a detailed understanding of bacterial metabolism: spectroscopic characterization of Staphylococcus epidermidis. ChemPhysChem. 2007;8(1):124–37.
Pahlow S, März A, Seise B, Hartmann K, Freitag I, Kämmer E, et al. Bioanalytical application of surface- and tip-enhanced Raman spectroscopy. Eng Life Sci. 2012;12(2):131–43.
Schmid T, Messmer A, Yeo B-S, Zhang W, Zenobi R. Towards chemical analysis of nanostructures in biofilms II: tip-enhanced Raman spectroscopy of alginates. Anal Bioanal Chem. 2008;391(5):1907–16.
Lyon LA, Keating CD, Fox AP, Baker BE, He L, Nicewarner SR, et al. Raman spectroscopy. Anal Chem. 1998;70(12):341R–61R.
Kano H, Segawa H, Okuno M, Leproux P, Couderc V. Hyperspectral coherent Raman imaging – principle, theory, instrumentation, and applications to life sciences. J Raman Spectrosc. 2016;47(1):116–23.
Krafft C, Popp J. The many facets of Raman spectroscopy for biomedical analysis. Anal Bioanal Chem. 2014;407(3):699–717.
Camp Jr CH, Cicerone MT. Chemically sensitive bioimaging with coherent Raman scattering. Nat Photonics. 2015;9(5):295–305.
Zhang X, Roeffaers MBJ, Basu S, Daniele JR, Fu D, Freudiger CW, et al. Label-free live-cell imaging of nucleic acids using stimulated Raman scattering microscopy. ChemPhysChem. 2012;13(4):1054–9.
Wei L, Yu Y, Shen Y, Wang MC, Min W. Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy. Proc Natl Acad Sci U S A. 2013;110(28):11226–31.
Petrov GI, Arora R, Yakovlev VV, Wang X, Sokolov AV, Scully MO. Comparison of coherent and spontaneous Raman microspectroscopies for noninvasive detection of single bacterial endospores. Proc Natl Acad Sci U S A. 2007;104(19):7776–9.
Wagner M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411–29.
Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9(9):683–94.
Wang Y, Huang WE, Cui L, Wagner M. Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr Opin Biotechnol. 2016;41:34–42.
Huang WE, Griffiths RI, Thompson IP, Bailey MJ, Whiteley AS. Raman microscopic analysis of single microbial cells. Anal Chem. 2004;76(15):4452–8.
Huang WE, Ward AD, Whiteley AS. Raman tweezers sorting of single microbial cells. Environ Microbiol Rep. 2009;1(1):44–9.
Wang Y, Ji Y, Wharfe ES, Meadows RS, March P, Goodacre R, et al. Raman activated cell ejection for isolation of single cells. Anal Chem. 2013;85(22):10697–701.
Muhamadali H, Chisanga M, Subaihi A, Goodacre R. Combining Raman and FT-IR spectroscopy with quantitative isotopic labeling for differentiation of E. coli cells at community and single cell levels. Anal Chem. 2015;87(8):4578–86.
Haider S, Wagner M, Schmid MC, Sixt BS, Christian JG, Häcker G, et al. Raman microspectroscopy reveals long-term extracellular activity of chlamydiae. Mol Microbiol. 2010;77(3):687–700.
Noothalapati H, Shigeto S. Exploring metabolic pathways in vivo by a combined approach of mixed stable isotope-labeled Raman microspectroscopy and multivariate curve resolution analysis. Anal Chem. 2014;86(15):7828–34.
Li M, Ashok PC, Dholakia K, Huang WE. Raman-activated cell counting for profiling carbon dioxide fixing microorganisms. J Phys Chem A. 2012;116(25):6560–3.
Kubryk P, Kölschbach JS, Marozava S, Lueders T, Meckenstock RU, Niessner R, et al. Exploring the potential of stable isotope (resonance) Raman microspectroscopy and surface-enhanced Raman scattering for the analysis of microorganisms at single cell level. Anal Chem. 2015;87(13):6622–30.
Eichorst SA, Strasser F, Woebken D, Woyke T, Schintlmeister A, Wagner M. Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol Ecol. 2015;91(10).
Berry D, Mader E, Lee TK, Woebken D, Wang Y, Zhu D, et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci U S A. 2015;112(2):E194–203.
Wang Y, Song Y, Tao Y, Muhamadali H, Goodacre R, Zhou N-Y, et al. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level. Anal Chem. 2016;88(19):9443–50.
Neufeld JD, Wagner M, Murrell JC. Who eats what, where and when? Isotope-labelling experiments are coming of age. ISME J. 2007;1(2):103–10.
Huang WE, Li M, Jarvis RM, Goodacre R, Banwart SA. Shining light on the microbial world: the application of Raman microspectroscopy. Adv Appl Microbiol. 2010;70:153–86.
Bocklitz T, Putsche M, Stüber C, Käs J, Niendorf A, Rösch P, et al. A comprehensive study of classification methods for medical diagnosis. J Raman Spectrosc. 2009;40(12):1759–65.
Stöckel S, Kirchhoff J, Neugebauer U, Rösch P, Popp J. The application of Raman spectroscopy for the detection and identification of microorganisms. J Raman Spectrosc. 2016;47(1):89–109.
Bocklitz T, Walter A, Hartmann K, Rösch P, Popp J. How to pre-process Raman spectra for reliable and stable models? Anal Chim Acta. 2011;704(1–2):47–56.
Stöckel S, Stanca AS, Helbig J, Rösch P, Popp J. Raman spectroscopic monitoring of the growth of pigmented and non-pigmented mycobacteria. Anal Bioanal Chem. 2015;407(29):8919–23.
Harz M, Rosch P, Peschke KD, Ronneberger O, Burkhardt H, Popp J. Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst. 2005;130(11):1543–50.
Maquelin K, Kirschner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, et al. Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Methods. 2002;51(3):255–71.
Acknowledgements
The authors gratefully acknowledge the German Research Foundation (Deutsche Forschungsgemeinschaft, project IV 110/2-1) and the Helmholtz Wasserzentrum München within the Helmholtz Water Network for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Ivleva, N.P., Kubryk, P. & Niessner, R. Raman microspectroscopy, surface-enhanced Raman scattering microspectroscopy, and stable-isotope Raman microspectroscopy for biofilm characterization. Anal Bioanal Chem 409, 4353–4375 (2017). https://doi.org/10.1007/s00216-017-0303-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0303-0