Abstract
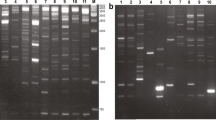
Allopolyploidization is considered an essential evolutionary process in plants that could trigger genomic shock in allopolyploid genome through activation of transcription of retrotransposons, which may be important in plant evolution. Two retrotransposon-based markers, inter-retrotransposon amplified polymorphism and retrotransposon-microsatellite amplified polymorphism and a microsatellite-based marker, inter simple sequence repeat were employed to investigate genomic changes in early generations of a newly synthesized allotetraploid Cucumis × hytivus Chen & Kirkbride (2n = 4x = 38) which was derived from crossing between cultivated cucumber C. sativus L. (2n = 2x = 14) and its wild relative C. hystrix Chakr. (2n = 2x = 24). Extensive genomic changes were observed, most of which involved the loss of parental DNA fragments and gain of novel fragments in the allotetraploid. Among the 28 fragments examined, 24 were lost while four were novel, suggesting that DNA sequence elimination is a relatively frequent event during polyploidization in Cucumis. Interestingly, of the 24 lost fragments, 18 were of C. hystrix origin, four were C. sativus-specific, and the remaining two were shared by both species, implying that fragment loss may be correlated with haploid DNA content (genome size) of diploid parents. Most changes were observed in the first generation after polyploidization (S1) and stably inherited in the subsequent three generations (S2–S4), indicating that genomic changes might be a rapid driving force for the stabilization of allotetraploids. Sequence analysis of 11 of the 28 altered DNA fragments showed that genomic changes in the allotetraploid occurred in both coding and non-coding regions, which might suggest that retrotransposons inserted into genome randomly and had a genome-wide effect on the allotetraploid evolution. Fluorescence in situ hybridization (FISH) analysis revealed a unique distribution of retrotransposon and/or microsatellite flanking sequences in mitotic and meiotic chromosomes, where the preferential FISH signals occurred in the centromeric and telomeric regions, implying that these regions were the possible hotspots for genomic changes.


Similar content being viewed by others
References
Adams KL, Cronn R, Percifeld R et al (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100:4649–4654
Baumel A, Ainouche M, Kalendar R et al (2002) Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Mol Biol Evol 19(8):1218–1227
Beaulieu J, Jean M, Belzile F (2009) The allotetraploid Arabidopsis thaliana-Arabidopsis lyrata subsp petraea as an alternative model system for the study of polyploidy in plants. Mol Genet Genomics 281:421–435
Bento M, Pereira HS, Rocheta M et al (2008) Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in Triticale. PLoS ONE 1(e1402):1–8
Bento M, Gustafson JP, Viegas W et al (2011) Size matters in Triticeae polyploids: larger genomes have higher remodeling. Genome 54:175–183
Chen LZ, Chen JF (2008) Changes of cytosine methylation induced by wide hybridization and allopolyploidy in Cucumis. Genome 51:789–799
Chen JF, Kirkbride JJ (2000) A new synthetic species of Cucumis (Cucurbitaceae) from interspecific hybridization and chromosome doubling. Brittonia 52:315–319
Chen JF, Staub JE, Tashiro Y et al (1997) Successful interspecific hybridization between Cucumis sativus L. and Cucumis hystrix Chakr. Euphytica 96:413–419
Chen LZ, Lou QF, Zhuang Y et al (2007) Cytological diploidization and rapid genome changes of the newly synthesized allotetraploids Cucumis × hytivus. Planta 225:603–614
Comai L (2000) Genetic and epigenetic interactions in allopolyploidy plants. Plant Mol Biol 43:387–399
Contento A, Heslop-Harrison JS, Schwarzacher T (2005) Diversity of a major repetitive DNA sequence in diploid and polyploid Triticaeae. Cytogenet Genome Res 109:34–42
Dernburg AF, Sedat JW, Hawley RS (1996) Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86:135–146
Dolezel J, Greihuber J, Lucretti S et al (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot 82(suppl 1):17–26
Feldman M, Levy AA (2005) Allopolyploidy-a shaping force in the evolution of wheat genomes. Cytogenet Genome Res 109:250–258
Feldman M, Liu B, Sehgal G et al (1997) Rapid elimination of low copy DNA sequence in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147:1381–1387
Han YH, Zhang ZH, Liu JH et al (2008) Distribution of the tandem repeat sequences and karyotyping in cucumber (Cucumis sativus L.) by fluorescence in situ hybridization. Cytogenet Genome Res 122:80–88
Jiang JM, Gill BS, Wang GL et al (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92:4487–4491
Jiang B, Lou QF, Wang D et al (2011) Allopolyploidization induced the activation of Ty1-copia retrotransposons in Cucumis hytivus, a newly formed Cucumis allotetraploid. Bot Stud 52:145–152
Josefsson C, Dilkes B, Comai L (2006) Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16:1322–1328
Kalendar R, Grob T, Regina M et al (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Karpen GH, Le MH, Le H (1996) Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 273:118–122
Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160:1651–1659
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kraitshtein Z, Yaakow B, Khasdan V et al (2010) Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186:801–812
Leitch IJ, Bennett MD (1997) Polyploidy in angiosperms. Trends Plant Sci 2:470–476
Leitch IJ, Bennett MD (2004) Genome downsizing in polyploid plants. Biol J Linn Soc 82:651–663
Liu B, Wendel JF (2000) Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43:874–880
Liu ZL, Wang YM, Shen Y et al (2004) Extensive alterations in DNA methylation and transcription in rice caused by introgression from Zizania latifolia. Plant Mol Biol 54:571–582
Ma XF, Gustafson JP (2006) Timing and rate of genome variation in triticale following allopolyploidization. Genome 49:950–958
Madlung A, Masuelli RW, Watson B et al (2002) Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allopolyploids. Plant Physiol 129:733–746
Madlung A, Tyagi AP, Watson B et al (2005) Genomic changes in synthetic Arabidopsis polyploids. Plant J 41:221–230
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Messing J, Bharti AK, Karlowski WM et al (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci USA 101:14349–14354
Murray HG, Thompson WF (1980) Rapid isolation of higher weight DNA. Nucl Acids Res 8:4321–4326
Ozkan H, Levy A, Feldman M (2001) Allopolyploid-induced rapid genomic evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13:1735–1747
Petit M, Guidat C, Daniel J et al (2010) Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytol 186:135–147
Shan XH, Liu ZL, Dong ZY et al (2005) Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol Biol Evol 22:976–990
Song K, Lu P, Tang K et al (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92:7719–7723
Volkov RA, Borisjuk NV, Panchuk II et al (1999) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol 16:311–320
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Zhuang Y, Chen JF (2009) Changes of gene expression in early generations of the synthetic allotetraploid Cucumis × hytivus Chen et Kirkbride. Genet Resour Crop Evol 56:1071–1076
Acknowledgments
This research was partially supported by the Key Program (30830079) and the General Program (31071801, 30972007) from the National Natural Science Foundation of China; National Basic Research Program of China (973 Program) (2009CB119000); the ‘863’ Programs (2008AA10Z150); the National Supporting Programs (2008BADB105) from the Ministry of Science and Technology of China; Ph. D. Funding (20070307034, 20090097110024) from the Ministry of Education of China; the Priority Academic Program Development of Jiangsu Higher Education Institution.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, B., Lou, Q., Wu, Z. et al. Retrotransposon- and microsatellite sequence-associated genomic changes in early generations of a newly synthesized allotetraploid Cucumis × hytivus Chen & Kirkbride. Plant Mol Biol 77, 225–233 (2011). https://doi.org/10.1007/s11103-011-9804-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9804-y