Abstract
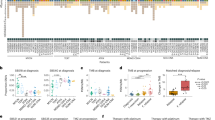
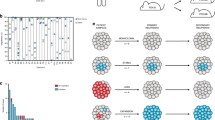
Medulloblastoma, the most common malignant paediatric brain tumour, arises in the cerebellum and disseminates through the cerebrospinal fluid in the leptomeningeal space to coat the brain and spinal cord1. Dissemination, a marker of poor prognosis, is found in up to 40% of children at diagnosis and in most children at the time of recurrence. Affected children therefore are treated with radiation to the entire developing brain and spinal cord, followed by high-dose chemotherapy, with the ensuing deleterious effects on the developing nervous system2. The mechanisms of dissemination through the cerebrospinal fluid are poorly studied, and medulloblastoma metastases have been assumed to be biologically similar to the primary tumour3,4. Here we show that in both mouse and human medulloblastoma, the metastases from an individual are extremely similar to each other but are divergent from the matched primary tumour. Clonal genetic events in the metastases can be demonstrated in a restricted subclone of the primary tumour, suggesting that only rare cells within the primary tumour have the ability to metastasize. Failure to account for the bicompartmental nature of metastatic medulloblastoma could be a major barrier to the development of effective targeted therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
CpG methylation data have been deposited in the Gene Expression Omnibus under accession number GSE34356. Reprints and permissions information is available at www.nature.com/reprints.
References
Gajjar, A. et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 7, 813–820 (2006)
Mabbott, D. J. et al. Serial evaluation of academic and behavioral outcome after treatment with cranial radiation in childhood. J. Clin. Oncol. 23, 2256–2263 (2005)
MacDonald, T. J. et al. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nature Genet. 29, 143–152 (2001)
Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. A molecular signature of metastasis in primary solid tumors. Nature Genet. 33, 49–54 (2003)
Goodrich, L. V., Milenkovic, L., Higgins, K. M. & Scott, M. P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997)
Collier, L. S., Carlson, C. M., Ravimohan, S., Dupuy, A. J. & Largaespada, D. A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276 (2005)
Dupuy, A. J., Akagi, K., Largaespada, D. A., Copeland, N. G. & Jenkins, N. A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005)
Helms, A. W., Abney, A. L., Ben-Arie, N., Zoghbi, H. Y. & Johnson, J. E. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185–1196 (2000)
Wetmore, C., Eberhart, D. E. & Curran, T. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res. 61, 513–516 (2001)
Brett, B. T. et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PLoS ONE 6, e24668 (2011)
Northcott, P. A. et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nature Genet. 41, 465–472 (2009)
Forbes, S. A. et al. in Current Protocols in Human Genetics Ch. 10.11. 10.1002/0471142905.hg1011s57 (2008)
Northcott, P. A. et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29, 1408–1414 (2011)
Nguyen, D. X. & Massague, J. Genetic determinants of cancer metastasis. Nature Rev. Genet. 8, 341–352 (2007)
Norton, L. & Massague, J. Is cancer a disease of self-seeding? Nature Med. 12, 875–878 (2006)
Luo, G., Ivics, Z., Izsvák, Z. & Bradley, A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 95, 10769–10773 (1998)
Rao, G., Pedone, C. A., Coffin, C. M., Holland, E. C. & Fults, D. W. c-Myc enhances sonic hedgehog-induced medulloblastoma formation from nestin-expressing neural progenitors in mice. Neoplasia 5, 198–204 (2003)
Swartling, F. J. et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 24, 1059–1072 (2010)
Pfister, S. et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J. Clin. Oncol. 27, 1627–1636 (2009)
Korshunov, A. et al. Accumulation of genomic aberrations during clinical progression of medulloblastoma. Acta Neuropathol. 116, 383–390 (2008)
Hahn, H. et al. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. J. Biol. Chem. 275, 28341–28344 (2000)
Fidler, I. J. & Kripke, M. L. Metastasis results from preexisting variant cells within a malignant tumor. Science 197, 893–895 (1977)
Scheel, C., Onder, T., Karnoub, A. & Weinberg, R. A. Adaptation versus selection: the origins of metastatic behavior. Cancer Res. 67, 11476–11480 (2007)
Talmadge, J. E. Clonal selection of metastasis within the life history of a tumor. Cancer Res. 67, 11471–11475 (2007)
Talmadge, J. E. & Fidler, I. J. The biology of cancer metastasis: historical perspective. Cancer Res. 70, 5649–5669 (2010)
Geurts, A. M. et al. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol. Ther. 8, 108–117 (2003)
Starr, T. K. et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323, 1747–1750 (2009)
Gnekow, A. K. Recommendations of the brain tumor subcommittee for the reporting of trials. Med. Pediatr. Oncol. 24, 104–108 (1995)
Acknowledgements
M.D.T. holds a Canadian Institutes of Health Research Clinician-Scientist Phase II Award, was a Sontag Foundation Distinguished Scholar, and is supported by grants from the National Institutes of Health (R01CA148699), the Pediatric Brain Tumor Foundation, the Canadian Cancer Society, and Brainchild. X.W. was supported by a fellowship from the American Brain Tumor Association in tribute to Tracy Greenwood. L.G. was supported by a fellowship from the Davis M. Ferguson Fund from the American Brain Tumor Association. A.D. was supported by a Vanier Doctoral Fellowship from the Canadian Institutes of Health Research. L.S.C. was supported by a grant (K01CA122183) and a Kimmel Scholar award from the Kimmel Foundation. C.E. was supported by a grant from the National Institutes of Health (NS055089). We thank S. Archer for technical writing assistance.
Author information
Authors and Affiliations
Contributions
M.D.T., X.W., P.A.N., L.S.C., A.D. and D.L. conceived the research and planned the experiments. X.W., P.A.N., A.D., L.G. and D.J.H.S. carried out the vast majority of the experiments under M.D.T.’s guidance. C.E., T.V.M., D.Z., Y.-J.C., T.M., X.-N.L., V.P.C., M.G.M. and W.A.W. provided human tumour materials. All authors contributed experimental expertise and participated in the writing of the manuscript. A.J.D., D.J.H.S., T.E.S., S.W.S., K.A., J.K., A.L.S., D.L. and L.S.C. provided biostatistical and bioinformatic expertise. E.B. provided the clinical data and analysis. D.W.F. carried out the Akt experiments. N.J., J.S. and J.M. carried out the exome sequencing. H.W., S.M.P. and A.K. carried out the immunostaining of human medulloblastoma tissue microarrays and FISH for MYCN and MYC. S.C. carried out the pathological analysis of mouse brain tumours.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-18 with legends. (PDF 7096 kb)
Supplementary Tables
This file contains Supplementary Tables 1-22. (PDF 20083 kb)
Rights and permissions
About this article
Cite this article
Wu, X., Northcott, P., Dubuc, A. et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 482, 529–533 (2012). https://doi.org/10.1038/nature10825
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10825
This article is cited by
-
Comparison of transcriptome profiles between medulloblastoma primary and recurrent tumors uncovers novel variance effects in relapses
Acta Neuropathologica Communications (2023)
-
Heterogeneity of the tumor immune microenvironment and clinical interventions
Frontiers of Medicine (2023)
-
An effective kinase inhibition strategy for metastatic recurrent childhood medulloblastoma
Journal of Neuro-Oncology (2023)
-
Primary leptomeningeal medulloblastoma: a case-based review
Child's Nervous System (2022)
-
Utilizing Carbon Ions to Treat Medulloblastomas that Exhibit Chromothripsis
Current Stem Cell Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.