Abstract
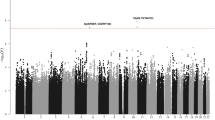
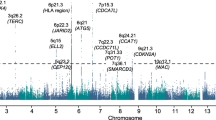
A number of specific chromosomal abnormalities define the subgroups of multiple myeloma. In a meta-analysis of two genome-wide association studies of multiple myeloma including a total of 1,661 affected individuals, we investigated risk for developing a specific tumor karyotype. The t(11;14)(q13;q32) translocation in which CCND1 is placed under the control of the immunoglobulin heavy chain enhancer was strongly associated with the CCND1 c.870G>A polymorphism (P = 7.96 × 10−11). These results provide a model in which a constitutive genetic factor is associated with risk of a specific chromosomal translocation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Anderson, K.C. & Carrasco, R.D. Pathogenesis of myeloma. Annu. Rev. Pathol. 6, 249–274 (2011).
Morgan, G.J., Walker, B.A. & Davies, F.E. The genetic architecture of multiple myeloma. Nat. Rev. Cancer 12, 335–348 (2012).
Sawyer, J.R. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 204, 3–12 (2011).
Broderick, P. et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat. Genet. 44, 58–61 (2012).
Power, C. & Elliott, J. Cohort profile: 1958 British birth cohort (National Child Development Study). Int. J. Epidemiol. 35, 34–41 (2006).
Schmermund, A. et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL Study. Risk Factors, Evaluation of Coronary Calcium and Lifestyle. Am. Heart J. 144, 212–218 (2002).
Bhattacharjee, S. et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am. J. Hum. Genet. 90, 821–835 (2012).
Jirawatnotai, S. et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature 474, 230–234 (2011).
Knudsen, K.E., Diehl, J.A., Haiman, C.A. & Knudsen, E.S. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene 25, 1620–1628 (2006).
Chng, W.J., Glebov, O., Bergsagel, P.L. & Kuehl, W.M. Genetic events in the pathogenesis of multiple myeloma. Best Pract. Res. Clin. Haematol. 20, 571–596 (2007).
Gostissa, M., Alt, F.W. & Chiarle, R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu. Rev. Immunol. 29, 319–350 (2011).
Fonseca, R. et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 64, 1546–1558 (2004).
Fernberg, P. et al. Genetic variation in chromosomal translocation breakpoint and immune function genes and risk of non-Hodgkin lymphoma. Cancer Causes Control 21, 759–769 (2010).
Pérez-Galán, P., Dreyling, M. & Wiestner, A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood 117, 26–38 (2011).
Pabalan, N. et al. Cyclin D1 Pro241Pro (CCND1-G870A) polymorphism is associated with increased cancer risk in human populations: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 17, 2773–2781 (2008).
Li, Z. et al. Alternate cyclin D1 mRNA splicing modulates p27KIP1 binding and cell migration. J. Biol. Chem. 283, 7007–7015 (2008).
Li, Z. et al. Alternative cyclin D1 splice forms differentially regulate the DNA damage response. Cancer Res. 70, 8802–8811 (2010).
Lévêque, C., Marsaud, V., Renoir, J.M. & Sola, B. Alternative cyclin D1 forms a and b have different biological functions in the cell cycle of B lymphocytes. Exp. Cell Res. 313, 2719–2729 (2007).
Marsaud, V. et al. Cyclin K and cyclin D1b are oncogenic in myeloma cells. Mol. Cancer 9, 103 (2010).
Morgan, G.J. et al. First-line treatment with zoledronic acid as compared with clodronic acid in multiple myeloma (MRC Myeloma IX): a randomised controlled trial. Lancet 376, 1989–1999 (2010).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Anderson, C.A. et al. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Pettiti, D. Meta-analysis, Decision Analysis and Cost-Effectiveness Analysis (Oxford University Press, New York, 1994).
Higgins, J.P. & Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Myers, S., Bottolo, L., Freeman, C., McVean, G. & Donnelly, P. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Chiecchio, L. et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia 20, 1610–1617 (2006).
Neben, K. et al. Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica 95, 1150–1157 (2010).
Walker, B.A. et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 108, 1733–1743 (2006).
Dimas, A.S. et al. Common regulatory variation impacts gene expression in a cell type–dependent manner. Science 325, 1246–1250 (2009).
Nica, A.C. et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 7, e1002003 (2011).
Stranger, B.E. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8, e1002639 (2012).
Stranger, B.E. et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1, e78 (2005).
Stranger, B.E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–853 (2007).
Slotta-Huspenina, J. et al. The impact of cyclin D1 mRNA isoforms, morphology and p53 in mantle cell lymphoma: p53 alterations and blastoid morphology are strong predictors of a high proliferation index. Haematologica 97, 1422–1430 (2012).
Acknowledgements
We are grateful to all the subjects and investigators at the individual centers for their participation. V.J. Forster is acknowledged for sample preparation. We also thank the staff of the CTRU (Clinical Trials Research Unit) University of Leeds and the NCRI (National Cancer Research Institute) Haematology Clinical Studies Group. In the UK, funding was provided by Myeloma UK, Leukaemia and Lymphoma Research (LLR 10021), Cancer Research UK (C1298/A8362, supported by the Bobby Moore Fund) and the National Health Service (NHS) via the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital NHS Trust. Leukaemia and Lymphoma Research provided funding for the Newcastle Myeloma Study (LLR 11006). In Germany, funding was provided by Dietmar-Hopp-Stiftung Walldorf, University Hospital Heidelberg and the German Cancer Aid. The HNR study was funded by the Heinz-Nixdorf Foundation (chairman M. Nixdorf) and the German Federal Ministry of Education and Research (BMBF).
Author information
Authors and Affiliations
Contributions
K.H., H.G., G.J.M. and R.S.H. designed the study. R.S.H. and G.J.M. obtained financial support in the UK, and K.H. and H.G. obtained financial support in Germany. R.S.H. and K.H. drafted the manuscript with contributions from G.J.M., D.C.J. and N.W. D.C., F.J.H., B.C., D.C.J., Y.P.M., S.E.D. and N.W. performed statistical and bioinformatics analyses. P.B. coordinated UK laboratory analyses. D.C.J. managed and prepared DNA samples for Myeloma IX and Myeloma XI Case Studies and performed expression analyses. H.G., K.N., N.W. and D.H. coordinated and managed German DNA samples. M.W.-H. and N.W. managed and prepared MCL samples. L.E. managed the HNR samples. P.H., T.W.M. and M.M.N. performed and coordinated genotyping of the German controls. K.H. and A.F. coordinated genotyping, performed by P.H. and M.M.N. D.C.J., M.F.K., N.L.L. and B.A.W. performed UK expression analyses. F.M.R. performed UK FISH analyses, and A.J. performed German FISH analyses. G.J.M., F.E.D., W.A.G. and G.H.J. performed ascertainment and collection of UK case samples. All authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 1–6 (PDF 3368 kb)
Rights and permissions
About this article
Cite this article
Weinhold, N., Johnson, D., Chubb, D. et al. The CCND1 c.870G>A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat Genet 45, 522–525 (2013). https://doi.org/10.1038/ng.2583
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2583
This article is cited by
-
PASTRY: achieving balanced power for detecting risk and protective minor alleles in meta-analysis of association studies with overlapping subjects
BMC Bioinformatics (2024)
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
Functional dissection of inherited non-coding variation influencing multiple myeloma risk
Nature Communications (2022)
-
A polygenic risk score for multiple myeloma risk prediction
European Journal of Human Genetics (2022)
-
Genetically determined telomere length and multiple myeloma risk and outcome
Blood Cancer Journal (2021)