Abstract
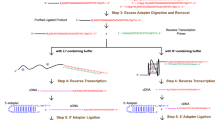
Most, if not all, known noncoding RNAs (ncRNAs) are associated with RNA binding proteins, thus forming ribonucleoprotein particles or RNPs. Here we describe a protocol for the generation of a specialized cDNA library from RNPs, thereby increasing the proportion of functional ncRNA species in the library. To that end, cellular extracts are fractionated on 10–30% glycerol gradients. Subsequently, RNP-derived ncRNAs are isolated and 3′-tailed by cytidine triphosphate and poly(A) polymerase; this is followed by 5′ adapter ligation by T4 RNA ligase. Reverse transcription of ncRNAs into cDNAs is carried out with an oligo-d(G) anchor primer. The generated cDNA libraries are subsequently submitted to high-throughput sequencing. This RNP selection procedure increases the probability of the presence of biologically relevant ncRNA species in the library compared with libraries generation methods that use size-selected, protein-devoid ncRNAs. The protocol enables the generation of deep-sequencing–compatible cDNA libraries that code for functional ncRNAs within 1 week.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Mattick, J.S. & Makunin, I.V. Small regulatory RNAs in mammals. Hum. Mol. Genet. 14 Spec No 1: R121–R132 (2005).
Willingham, A.T. & Gingeras, T.R. TUF love for 'junk' DNA. Cell 125, 1215–1220 (2006).
Huttenhofer, A., Schattner, P. & Polacek, N. Non-coding RNAs: hope or hype? Trends Genet. 21, 289–297 (2005).
Brosius, J. Waste not, want not—transcript excess in multicellular eukaryotes. Trends Genet. 21, 287–288 (2005).
Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2, 919–929 (2001).
Huttenhofer, A. & Schattner, P. The principles of guiding by RNA: chimeric RNA-protein enzymes. Nat. Rev. Genet. 7, 475–482 (2006).
Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Bachellerie, J.P., Cavaille, J. & Huttenhofer, A. The expanding snoRNA world. Biochimie 84, 775–790 (2002).
Rederstorff, M. et al. RNPomics: defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 38, e113 (2010).
Huttenhofer, A., Brosius, J. & Bachellerie, J.P. RNomics: identification and function of small, non-messenger RNAs. Curr. Opin. Chem. Biol. 6, 835–843 (2002).
Huttenhofer, A., Cavaille, J. & Bachellerie, J.P. Experimental RNomics: a global approach to identifying small nuclear RNAs and their targets in different model organisms. Methods Mol. Biol. 265, 409–428 (2004).
Huttenhofer, A. & Vogel, J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 34, 635–646 (2006).
Jochl, C. et al. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 36, 2677–2689 (2008).
Lung, B. et al. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 34, 3842–3852 (2006).
Madej, M.J., Alfonzo, J.D. & Huttenhofer, A. Small ncRNA transcriptome analysis from kinetoplast mitochondria of Leishmania tarentolae. Nucleic Acids Res. 35, 1544–1554 (2007).
Mrazek, J., Kreutmayer, S.B., Grasser, F.A., Polacek, N. & Huttenhofer, A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 35, e73 (2007).
Mercer, T.R., Dinger, M.E. & Mattick, J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 (2009).
Kapranov, P. et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 15, 987–997 (2005).
Marioni, J.C., Mason, C.E., Mane, S.M., Stephens, M. & Gilad, Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517 (2008).
Wang, Z., Gerstein, M. & Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
Velculescu, V.E., Zhang, L., Vogelstein, B. & Kinzler, K.W. Serial analysis of gene expression. Science 270, 484–487 (1995).
Fullwood, M.J., Wei, C.L., Liu, E.T. & Ruan, Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 19, 521–532 (2009).
Dignam, J.D., Lebovitz, R.M. & Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Huttenhofer, A. et al. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO. J. 20, 2943–2953 (2001).
Acknowledgements
This work was supported by the Medical University of Innsbruck and the Austrian Genome Research (GEN-AU; http://www.gen-au.at/) funding program (grants D-110420-011-013 and D-11420-011-015 to A.H.). We acknowledge M. Lukasser, K. Skreka and M. Erlacher for technical assistance.
Author information
Authors and Affiliations
Contributions
M.R. designed and performed the experiments and wrote the manuscript. A.H. designed the experiments and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Rederstorff, M., Hüttenhofer, A. cDNA library generation from ribonucleoprotein particles. Nat Protoc 6, 166–174 (2011). https://doi.org/10.1038/nprot.2010.186
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.186
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.