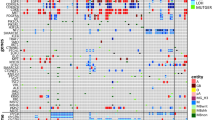
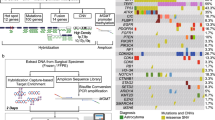
Abstract
With the number of prognostic and predictive genetic markers in neuro-oncology steadily growing, the need for comprehensive molecular analysis of neuropathology samples has vastly increased. We therefore developed a customized enrichment/hybrid-capture-based next-generation sequencing (NGS) gene panel comprising the entire coding and selected intronic and promoter regions of 130 genes recurrently altered in brain tumors, allowing for the detection of single nucleotide variations, fusions, and copy number aberrations. Optimization of probe design, library generation and sequencing conditions on 150 samples resulted in a 5-workday routine workflow from the formalin-fixed paraffin-embedded sample to neuropathological report. This protocol was applied to 79 retrospective cases with established molecular aberrations for validation and 71 prospective cases for discovery of potential therapeutic targets. Concordance of NGS compared to established, single biomarker methods was 98.0 %, with discrepancies resulting from one case where a TERT promoter mutation was not called by NGS and three ATRX mutations not being detected by Sanger sequencing. Importantly, in samples with low tumor cell content, NGS was able to identify mutant alleles that were not detectable by traditional methods. Information derived from NGS data identified potential targets for experimental therapy in 37/47 (79 %) glioblastomas, 9/10 (90 %) pilocytic astrocytomas, and 5/14 (36 %) medulloblastomas in the prospective target discovery cohort. In conclusion, we present the settings for high-throughput, adaptive next-generation sequencing in routine neuropathology diagnostics. Such an approach will likely become highly valuable in the near future for treatment decision making, as more therapeutic targets emerge and genetic information enters the classification of brain tumors.


Similar content being viewed by others
References
BCL to fastq conversion. https://www.support.illumina.com/content/dam/illumina-support/documents/documentation/software_documentation/bcl2fastq/bcl2fastq2-v216-Software-Guide-15051736-E.pdf. Accessed 8 Oct 2015
Fastqc. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 8 Oct 2015
Picard duplicate removal. http://broadinstitute.github.io/picard/. Accessed 8 Oct 2015
SAMtools. https://samtools.github.io/hts-specs/SAMv1.pdf. Accessed 8 Oct 2015
SAMtools mpileup. http://www.samtools.sourceforge.net/mpileup.shtml. Accessed 8 Oct 2015
SAMtools vcf. http://samtools.github.io/hts-specs/VCFv4.2.pdf. Accessed 8 Oct 2015
Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G et al (2011) Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol 122(1):11–19
Choi BD, Archer GE, Mitchell DA, Heimberger AB, McLendon RE, Bigner DD et al (2009) EGFRvIII-targeted vaccination therapy of malignant glioma. Brain Pathol 19(4):713–723
Cimino PJ, Bredemeyer A, Abel HJ, Duncavage EJ (2015) A wide spectrum of EGFR mutations in glioblastoma is detected by a single clinical oncology targeted next-generation sequencing panel. Exp Mol Pathol 98(3):568–573
Do H, Dobrovic A (2015) Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem 61(1):64–71
Do H, Wong SQ, Li J, Dobrovic A (2013) Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clin Chem 59(9):1376–1383
Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD et al (2015) Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 17(4):385–399
Hovestadt V, Zapatka M. Enhanced copy-number variation analysis using Illumina 450 k methylation arrays. https://www.bioconductor.org/packages/release/bioc/html/conumee.html. Accessed 8 Oct 2015
Hummel M, Bonnin S, Lowy E, Roma G (2011) TEQC: an R package for quality control in target capture experiments. Bioinformatics 27(9):1316–1317
Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ et al (2013) Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 45(8):927–932
Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V et al (2014) Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25(3):393–405
Korshunov A, Meyer J, Capper D, Christians A, Remke M, Witt H et al (2009) Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol 118(3):401–405
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595
Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A et al (2014) International Society Of Neuropathology-Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 24(5):429–435
Louis DN, Virgin HWT, Asa SL (2011) “Next-generation” pathology and laboratory medicine. Arch Pathol Lab Med 135(12):1531–1532
Malapelle U, Vigliar E, Sgariglia R, Bellevicine C, Colarossi L, Vitale D et al (2015) Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. J Clin Pathol 68(1):64–68
Manson-Bahr D, Ball R, Gundem G, Sethia K, Mills R, Rochester M et al (2015) Mutation detection in formalin-fixed prostate cancer biopsies taken at the time of diagnosis using next-generation DNA sequencing. J Clin Pathol 68(3):212–217
McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG et al (2011) deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7(5):e1001138
Mosen-Ansorena D, Telleria N, Veganzones S, De la Orden V, Maestro ML, Aransay AM (2014) seqCNA: an R package for DNA copy number analysis in cancer using high-throughput sequencing. BMC Genom 15:178
Obenchain V, Lawrence M, Carey V, Gogarten S, Shannon P, Morgan M (2014) VariantAnnotation: a bioconductor package for exploration and annotation of genetic variants. Bioinformatics 30(14):2076–2078
Reuss DE, Kratz A, Sahm F, Capper D, Schrimpf D, Koelsche C et al (2015) Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol 130(3):407–417
Reuss DE, Mamatjan Y, Schrimpf D, Capper D, Hovestadt V, Kratz A et al (2015) IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol 129(6):867–873
Reuss DE, Sahm F, Schrimpf D, Wiestler B, Capper D, Koelsche C et al (2015) ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol 129(1):133–146
Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SR, Consortium WGS et al (2014) Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 46(8):912–918
Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, Quandt J et al (2014) A vaccine targeting mutant IDH1 induces antitumour immunity. Nature 512(7514):324–327
Schweizer L, Koelsche C, Sahm F, Piro RM, Capper D, Reuss DE et al (2013) Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein. Acta Neuropathol 125(5):651–658
Serizawa M, Yokota T, Hosokawa A, Kusafuka K, Sugiyama T, Tsubosa Y et al (2015) The efficacy of uracil DNA glycosylase pretreatment in amplicon-based massively parallel sequencing with DNA extracted from archived formalin-fixed paraffin-embedded esophageal cancer tissues. Cancer Genet 208(9):415–427
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22(4):425–437
Suva ML, Louis DN (2013) Next-generation molecular genetics of brain tumours. Curr Opin Neurol 26(6):681–687
Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR et al (2010) Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn JMD 12(4):425–432
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acid Res 38(16):e164
Acknowledgments
This study was supported by the German Cancer Aid (110670) and by a PostDoc fellowship of the Medical Faculty Heidelberg to FS. We also thank the DKFZ-Heidelberg Center for Personalized Oncology (DKFZ-HIPO) for funding through HIPO_036 and HIPO_057.
Author information
Authors and Affiliations
Corresponding author
Additional information
F. Sahm, D. Schrimpf, D. T. W. Jones and J. Meyer contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sahm, F., Schrimpf, D., Jones, D.T.W. et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol 131, 903–910 (2016). https://doi.org/10.1007/s00401-015-1519-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-015-1519-8